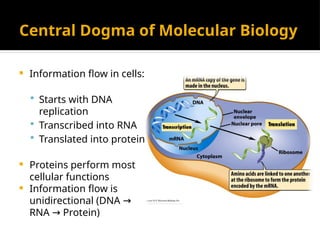
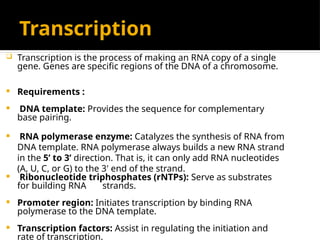
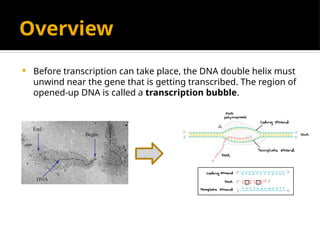
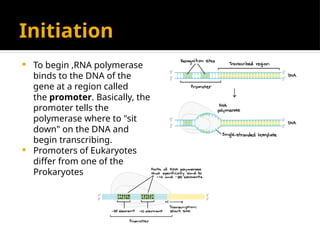
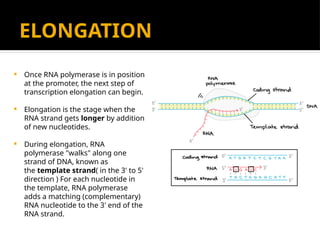
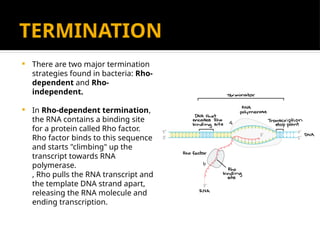
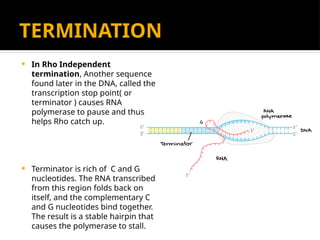
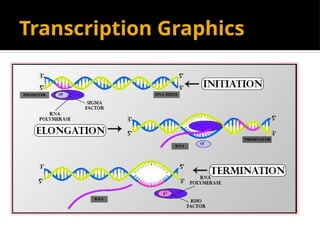
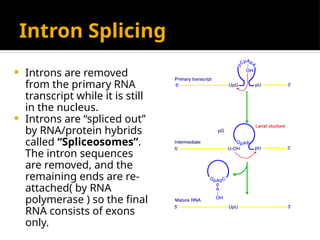
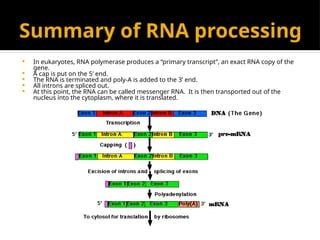
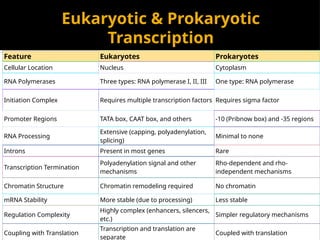
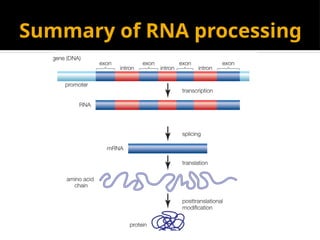
The document discusses transcription as a key process in the central dogma of molecular biology, detailing the transformation of DNA into RNA via RNA polymerase, which progresses through initiation, elongation, and termination steps. It highlights the difference between prokaryotic and eukaryotic transcription, focusing on RNA processing in eukaryotes, such as capping, polyadenylation, and intron splicing. The document also compares the roles of RNA polymerase and the complexity of transcription regulation in both cell types.