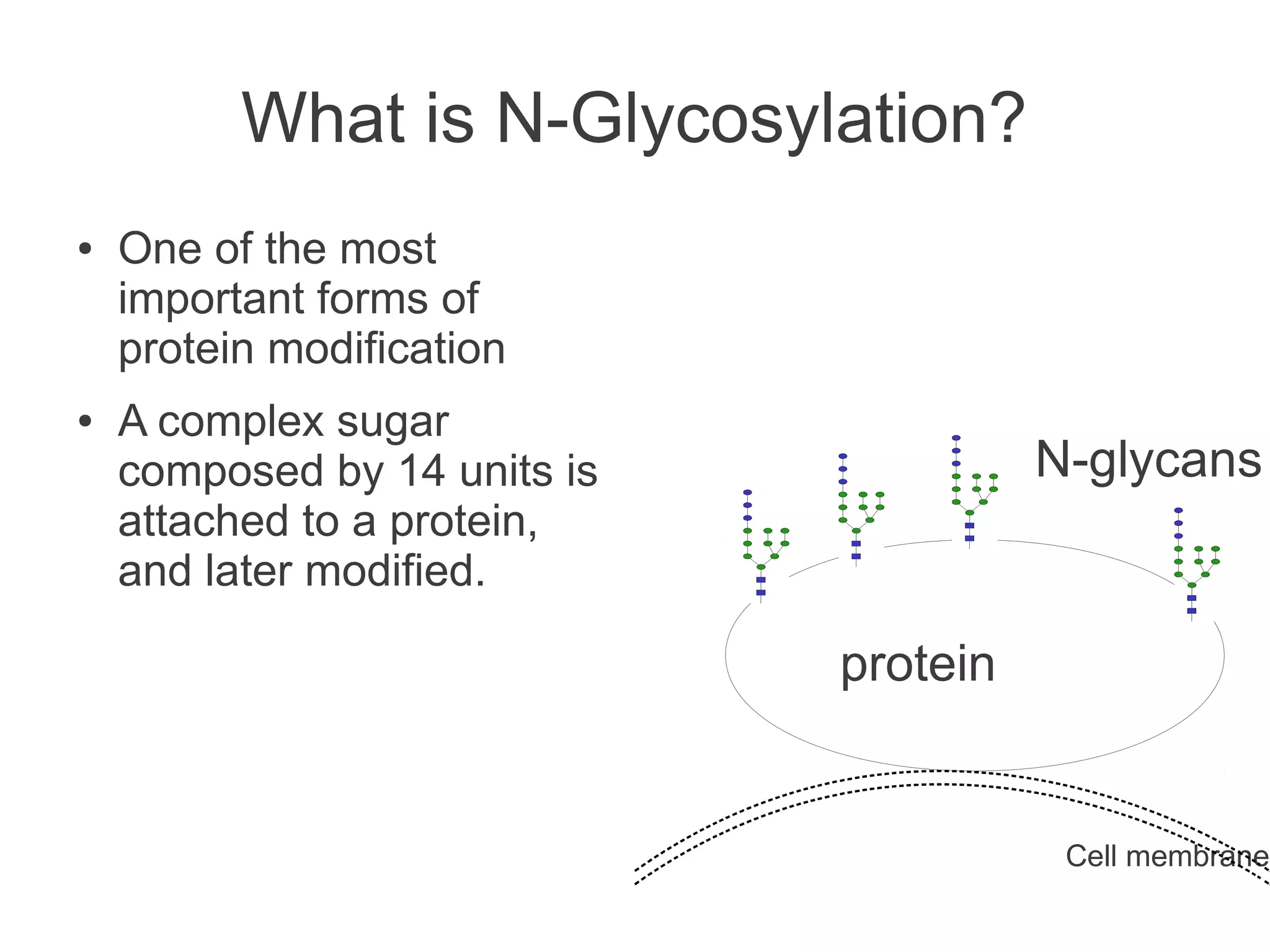
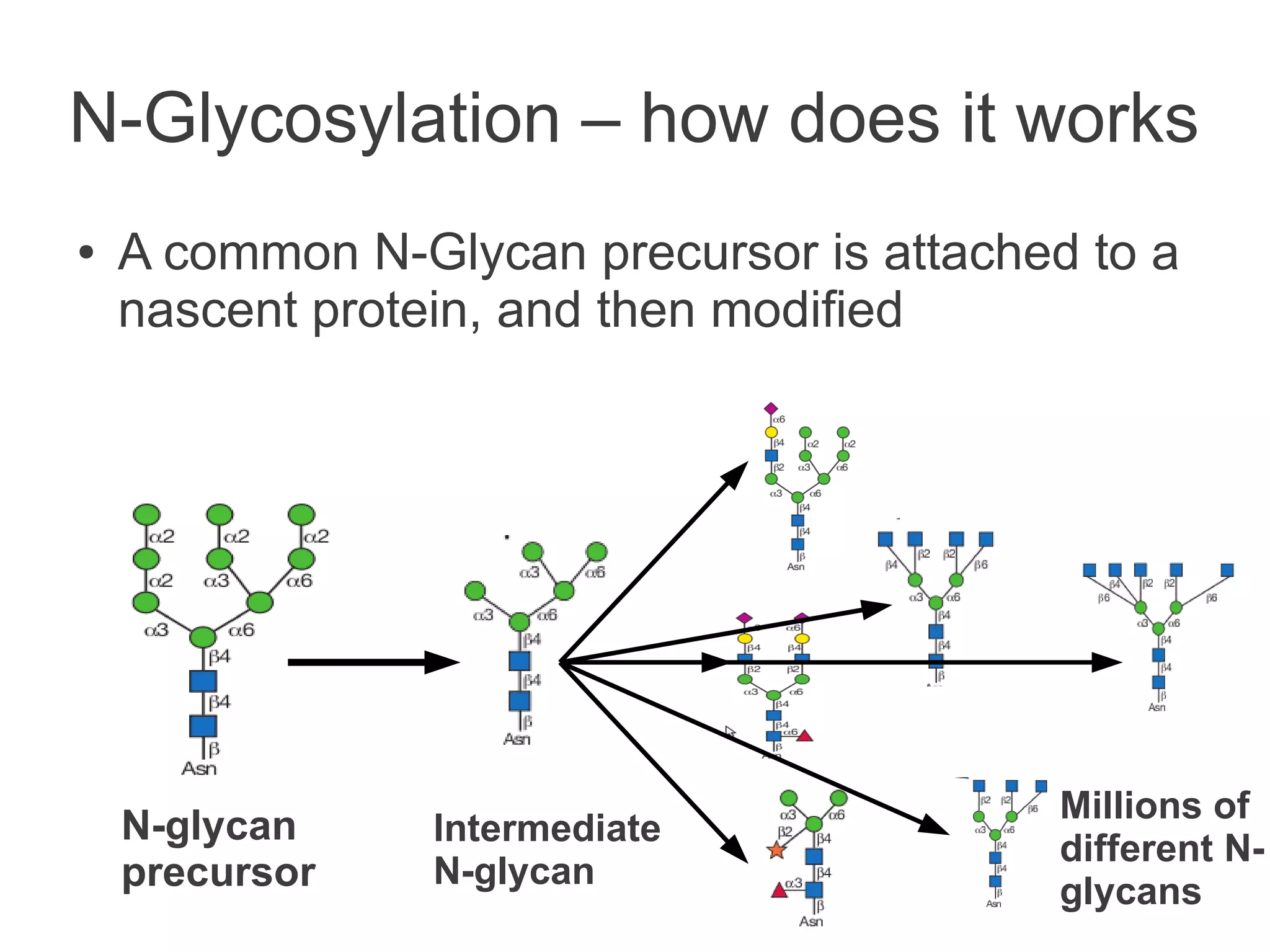
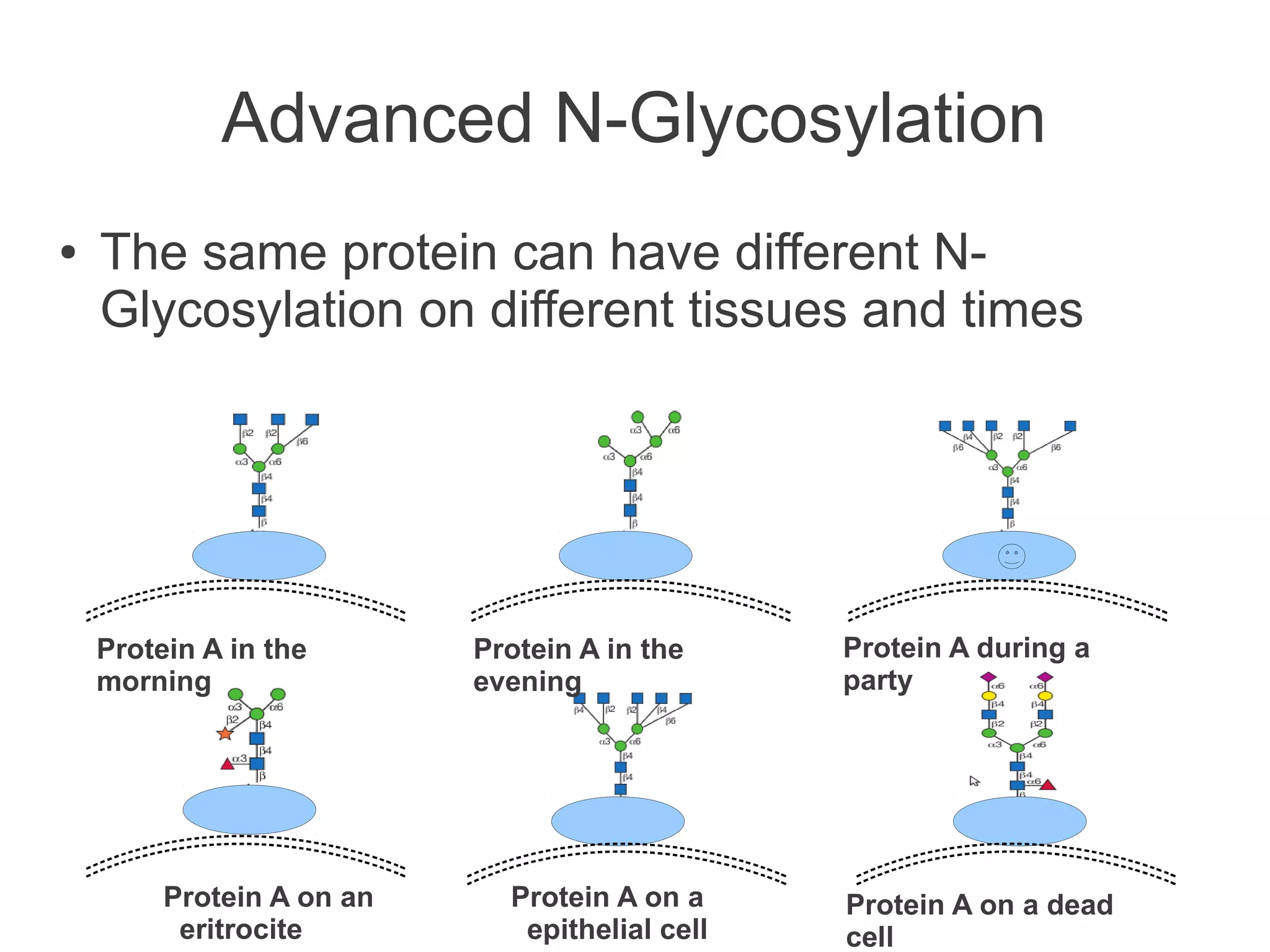
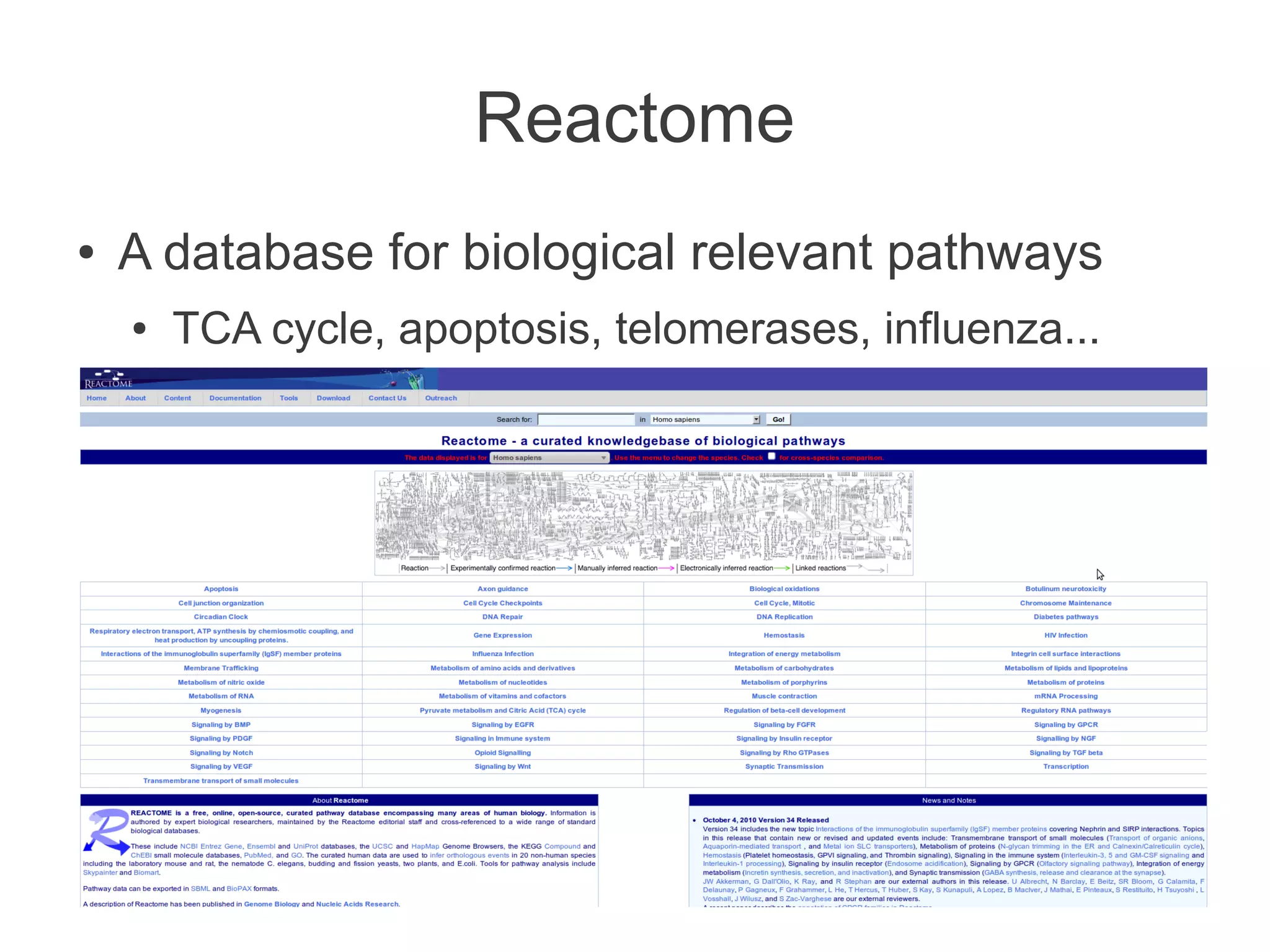
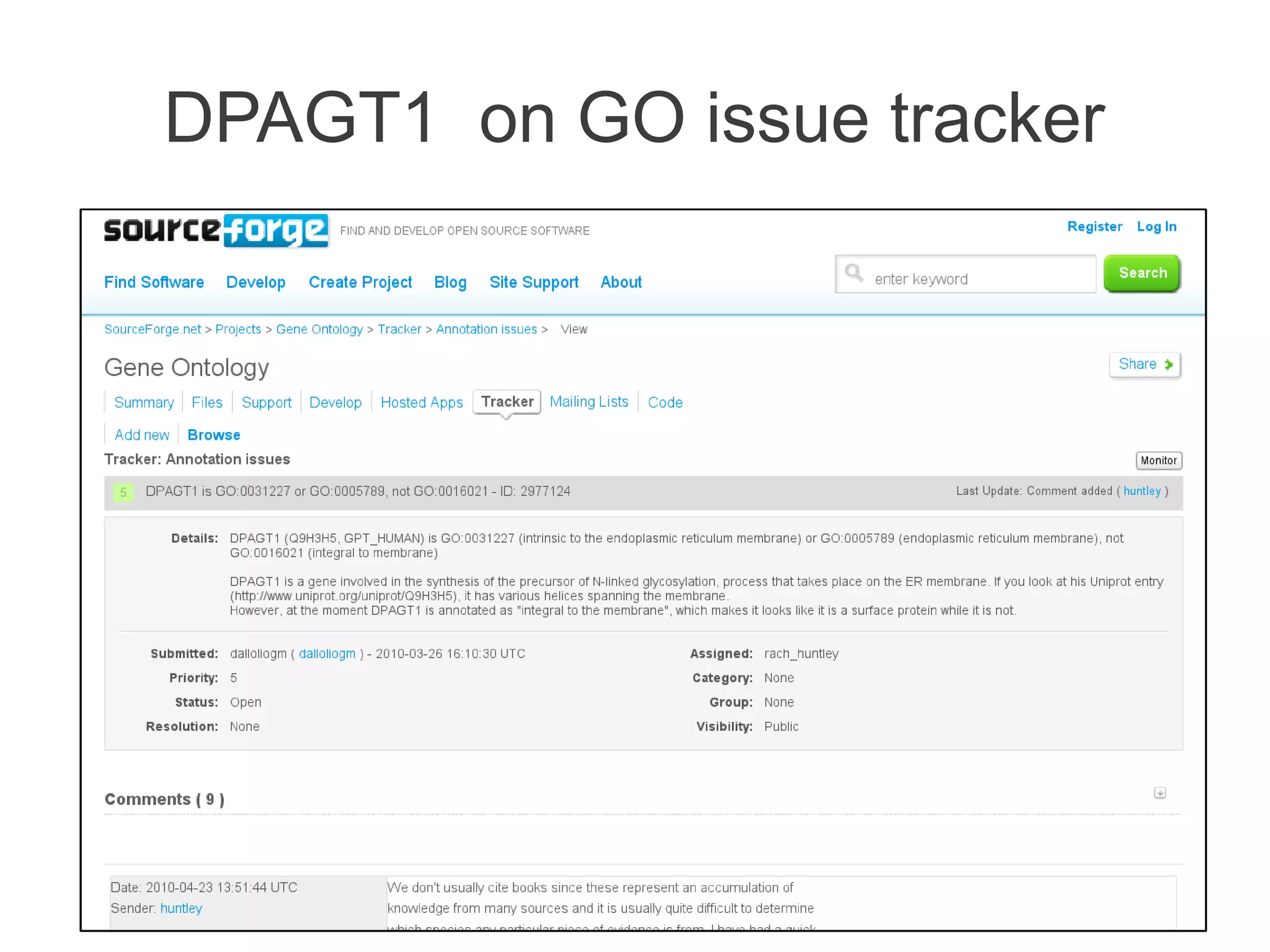
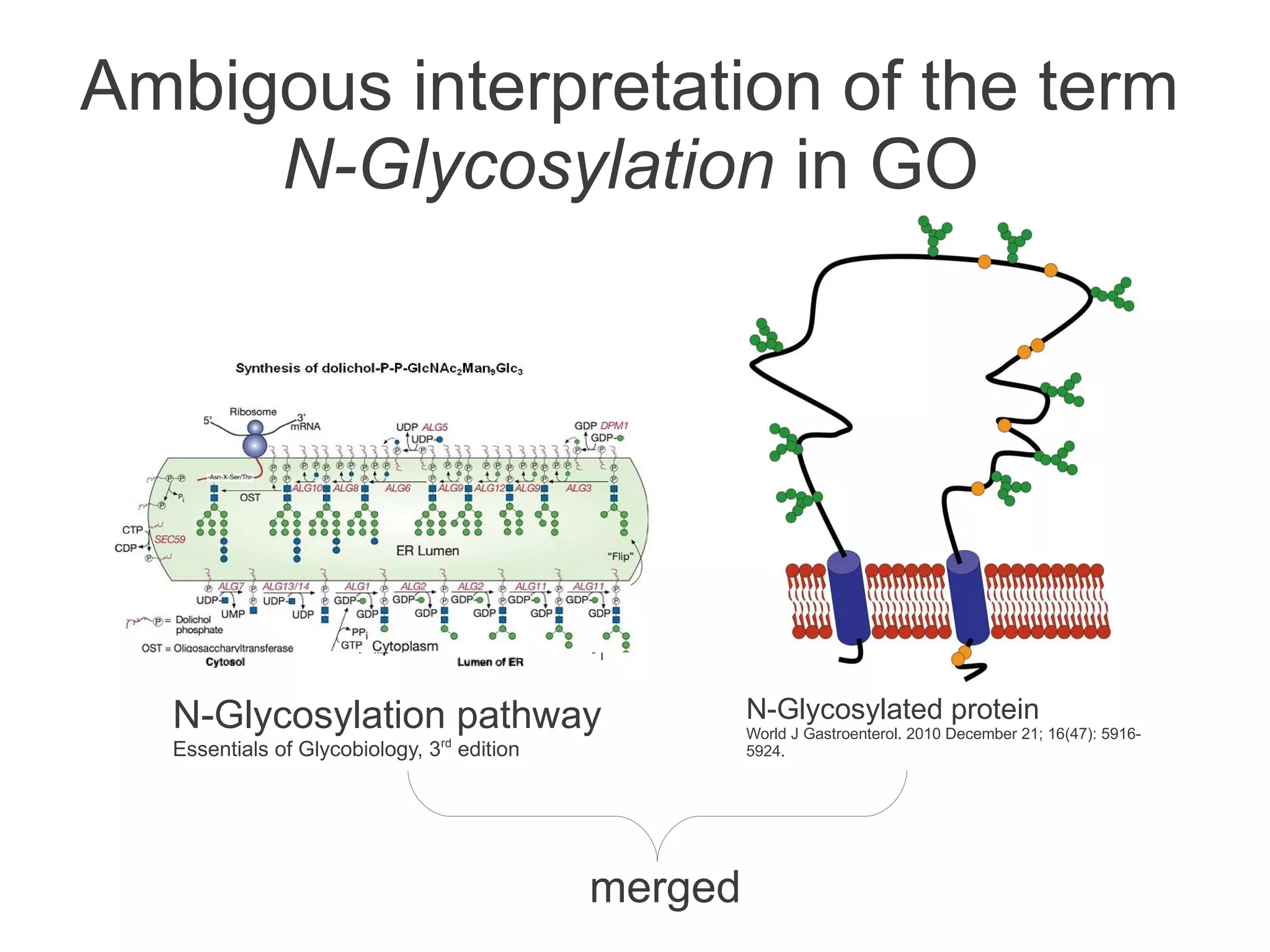
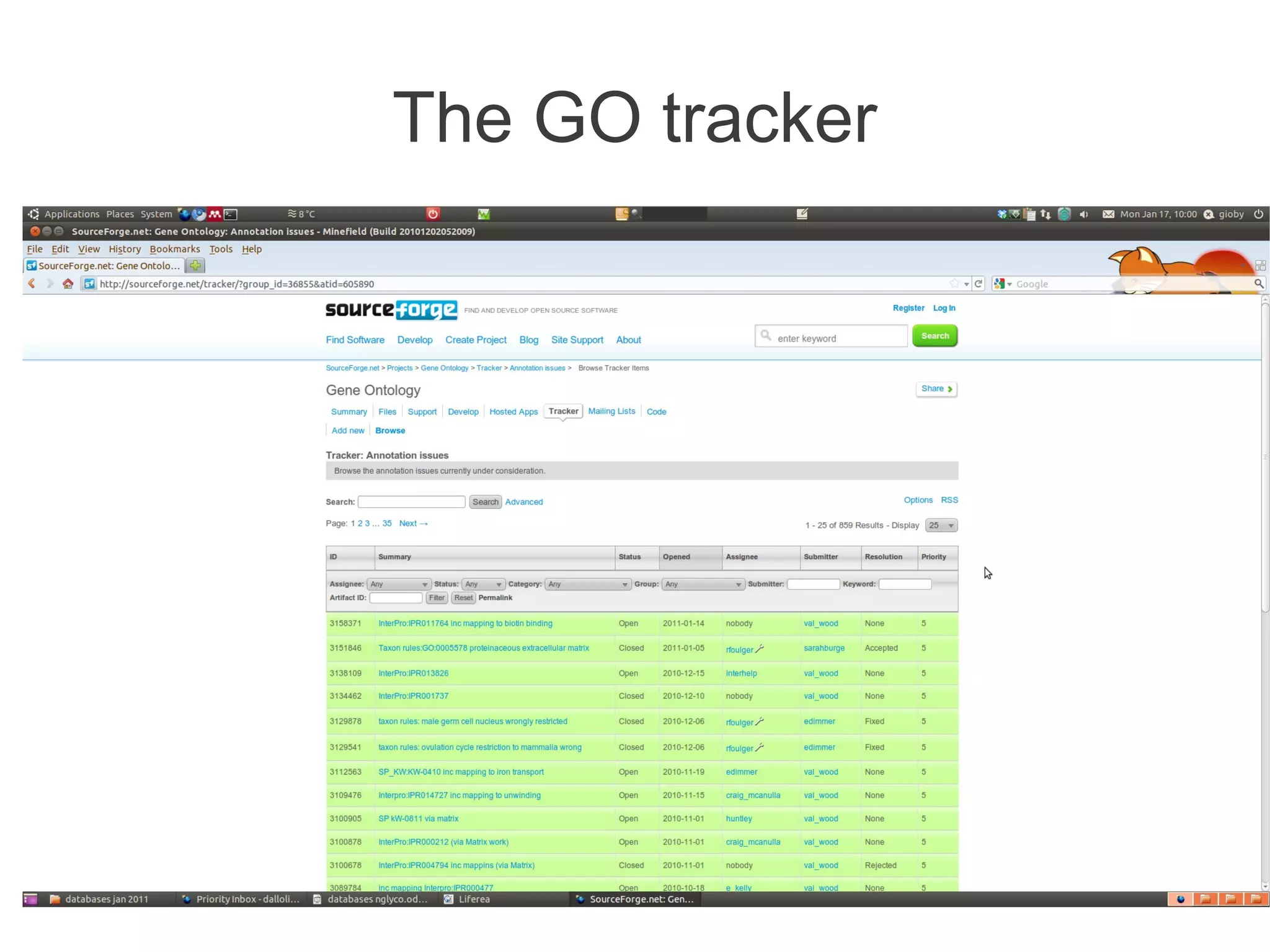
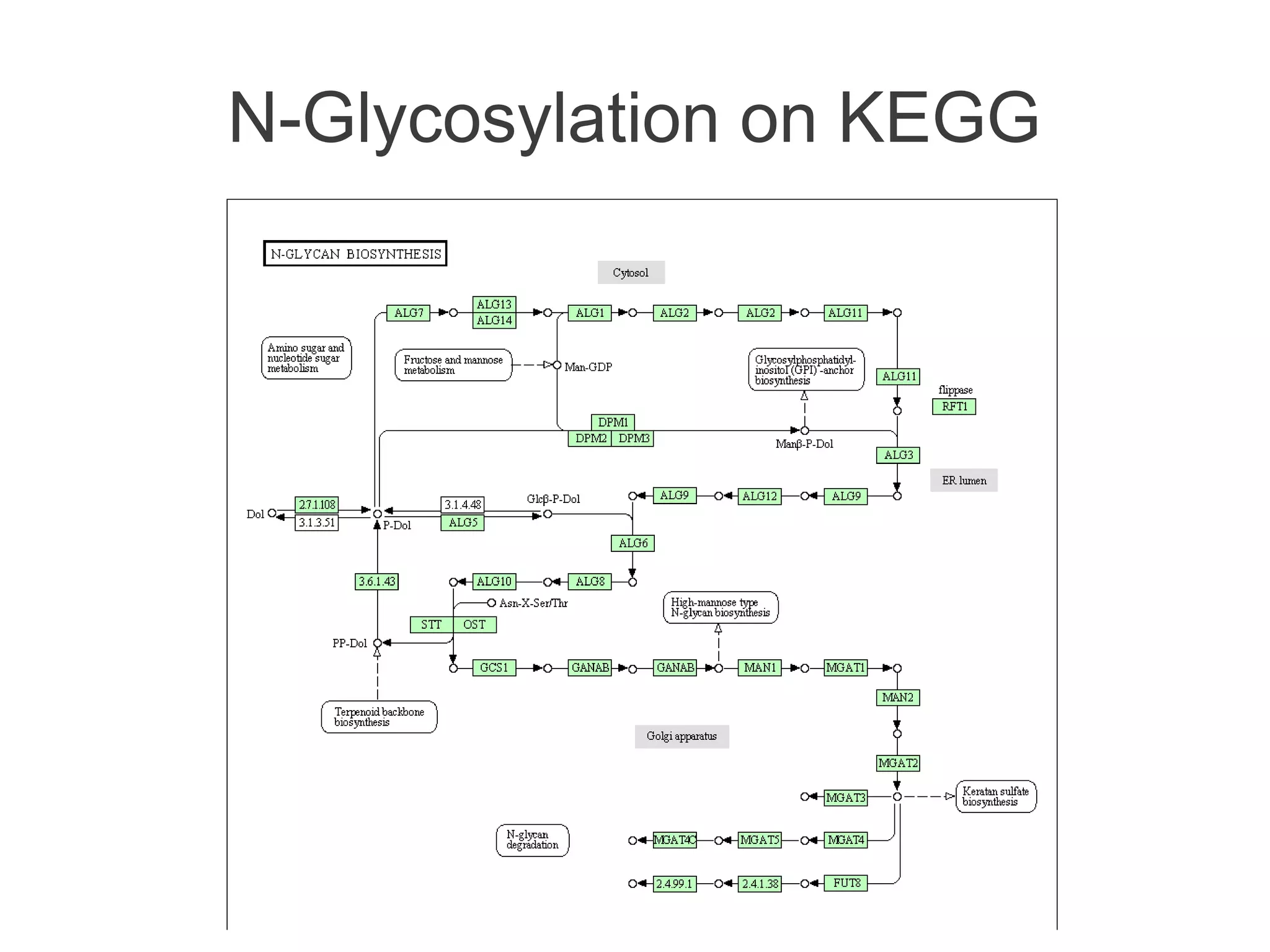
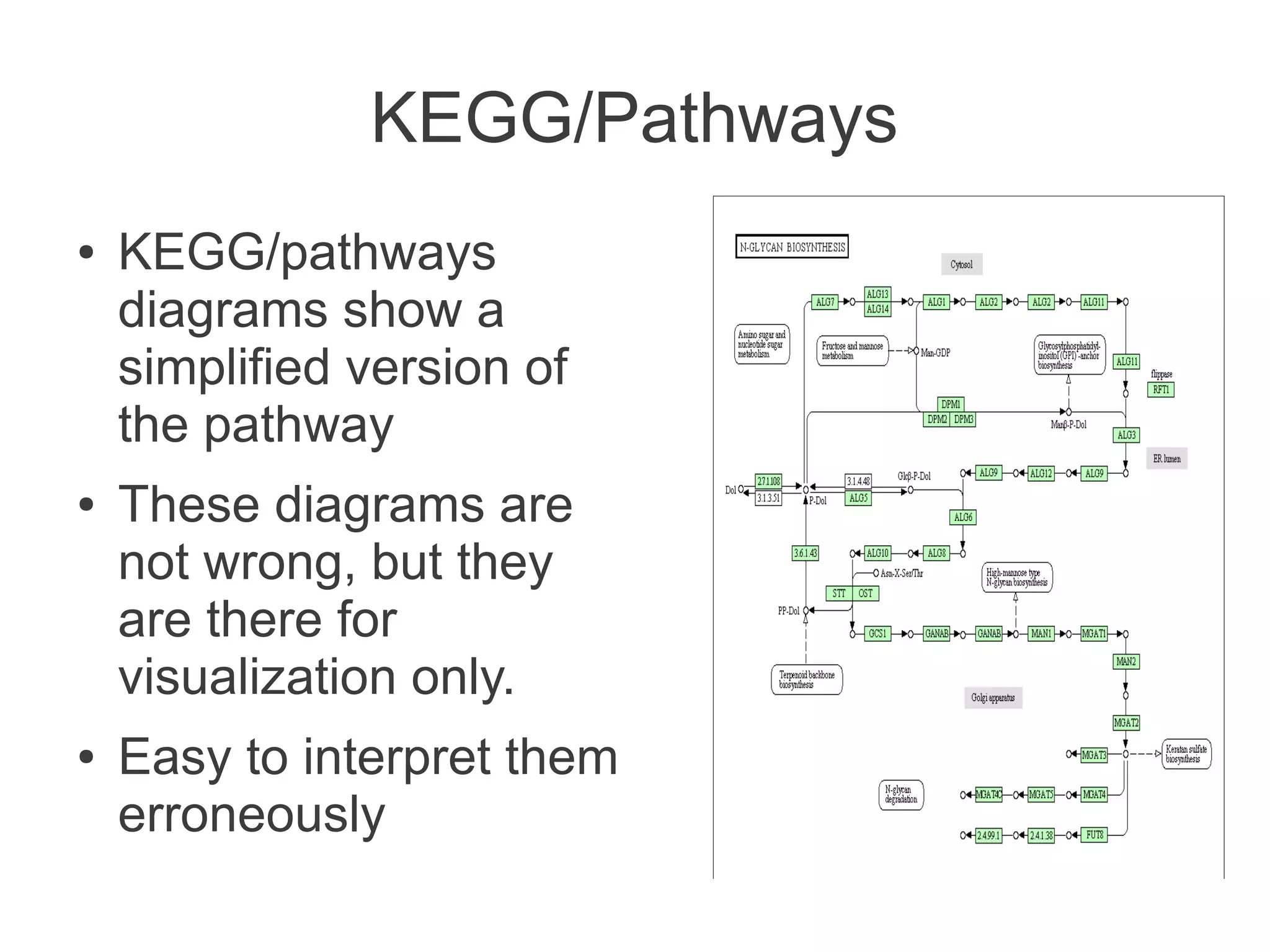
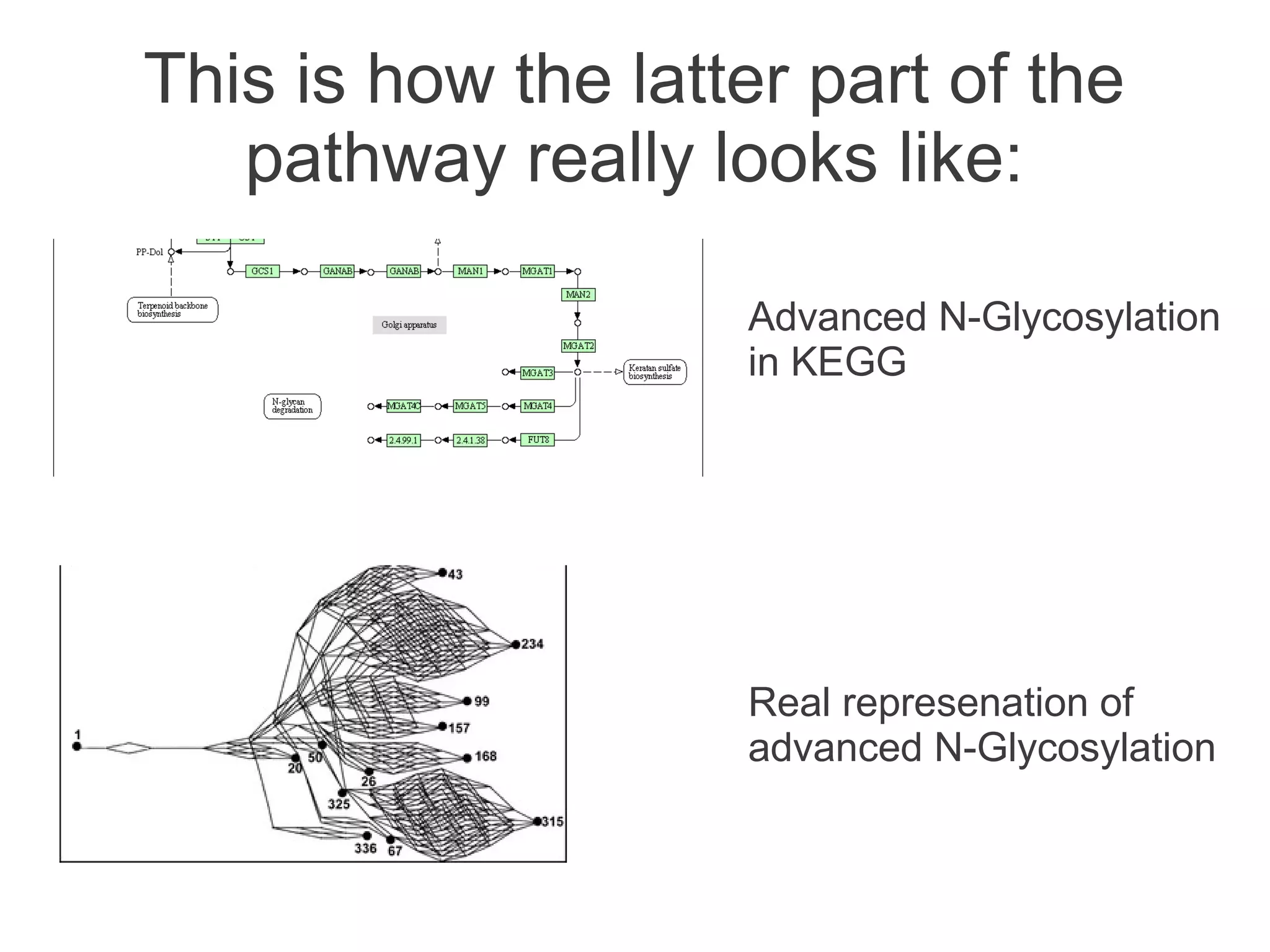
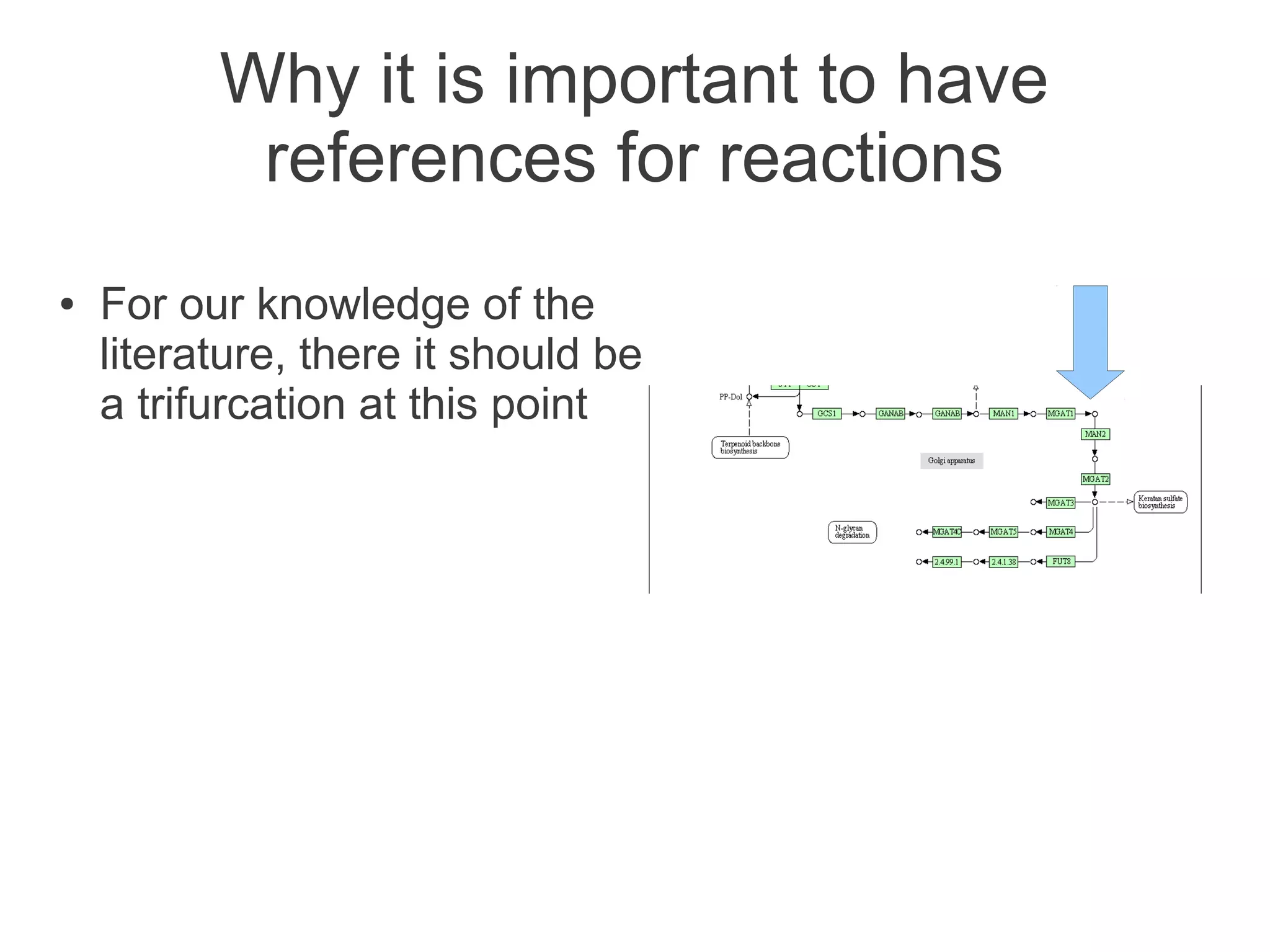
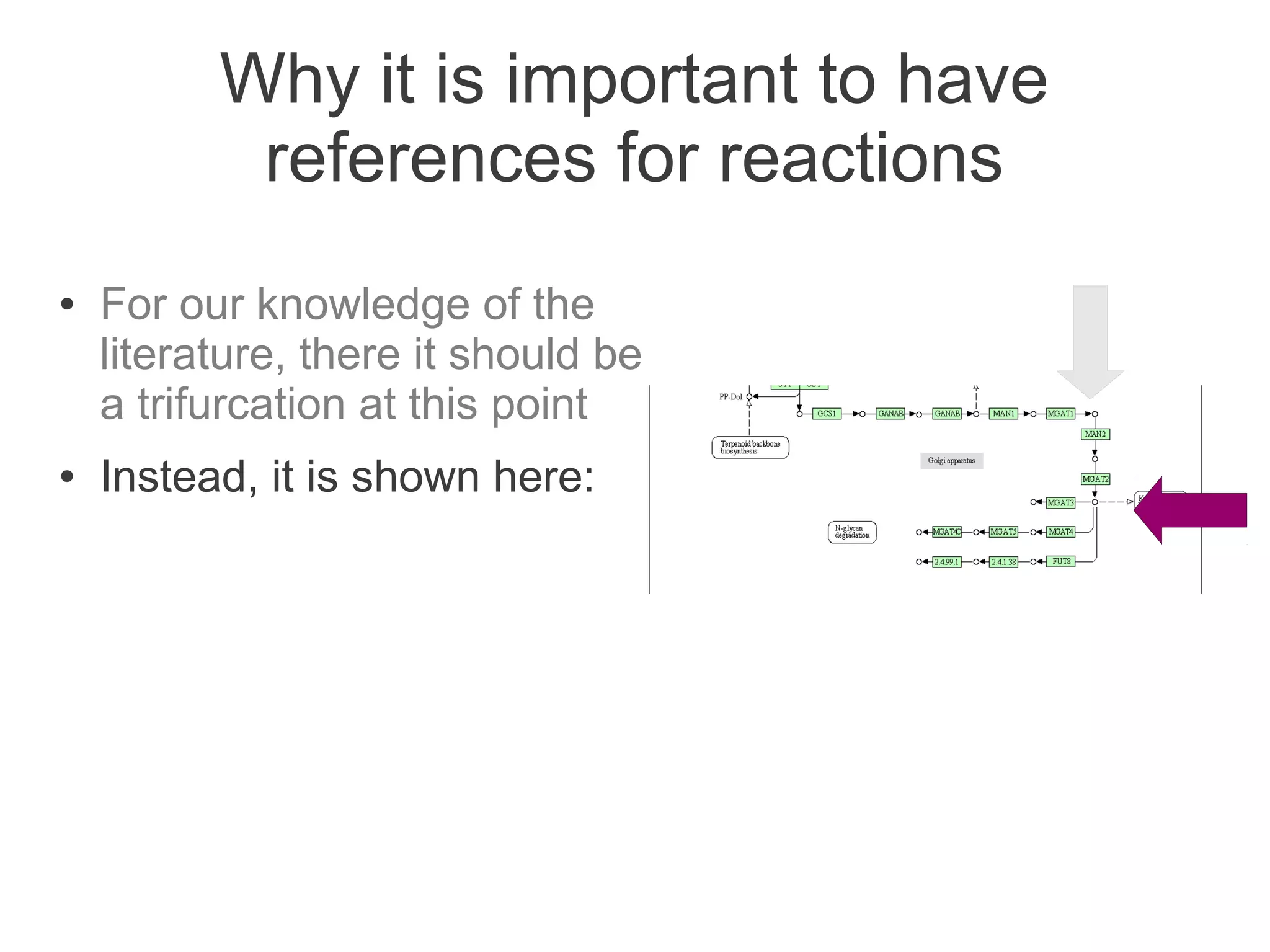
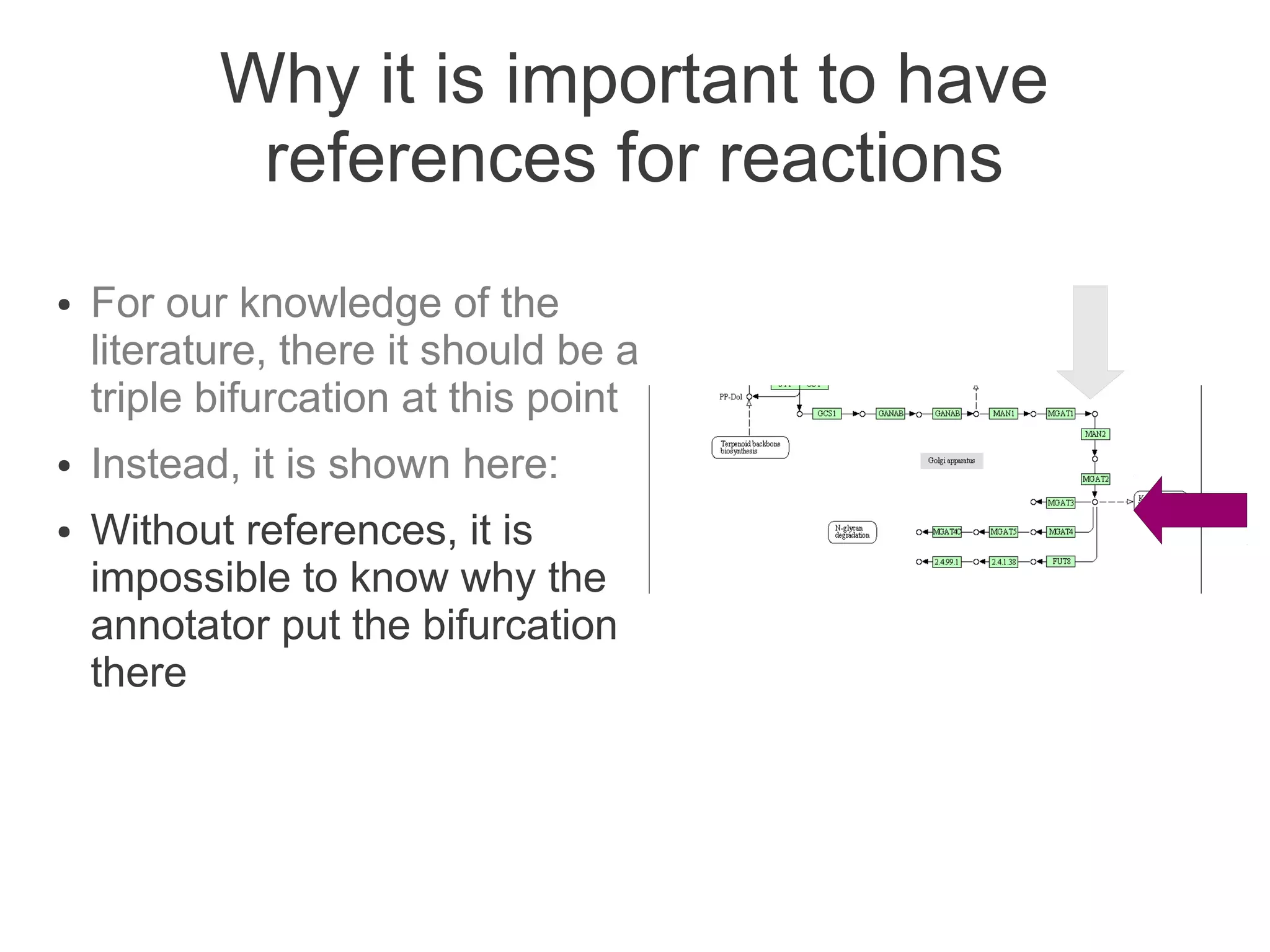
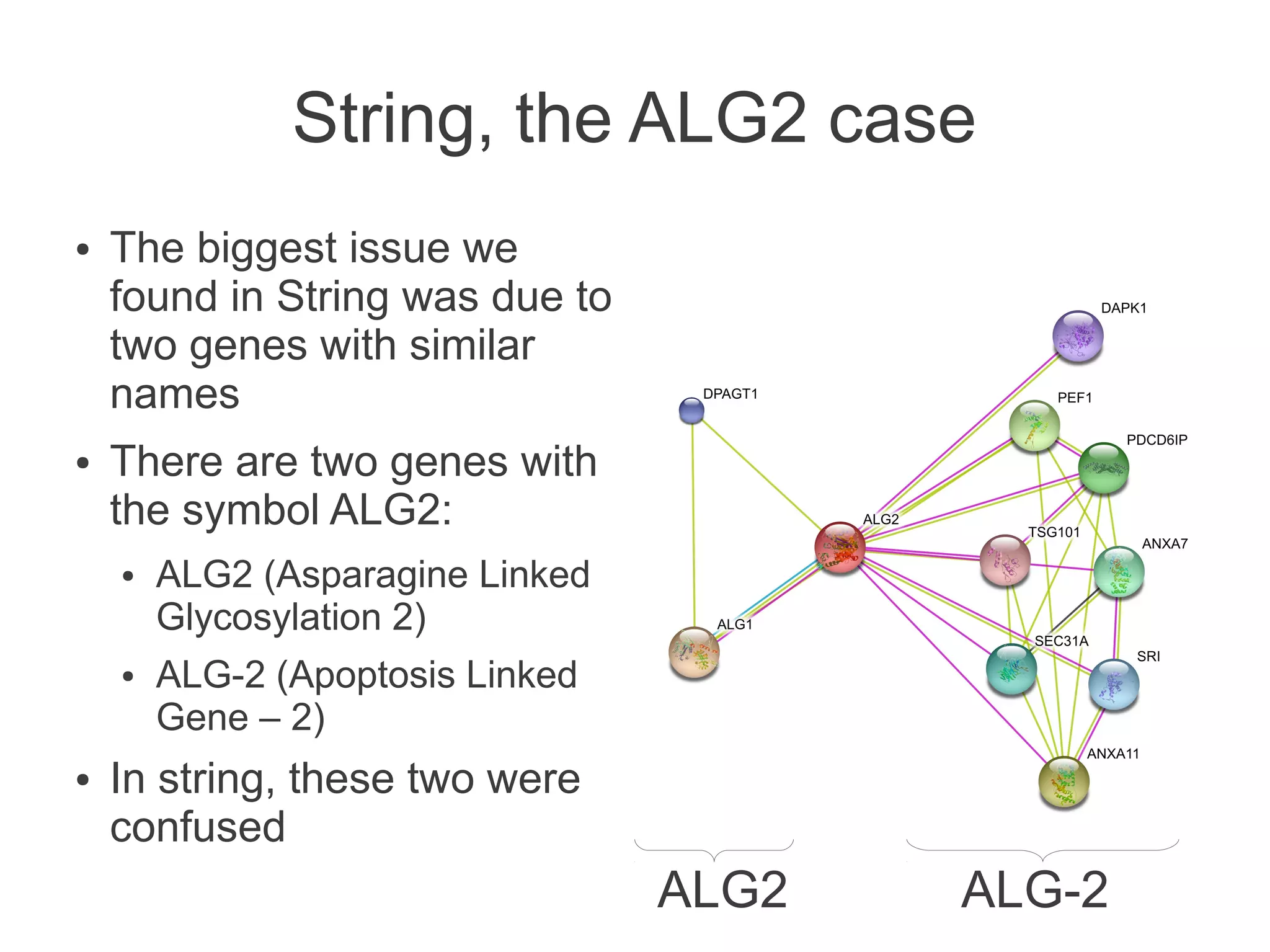
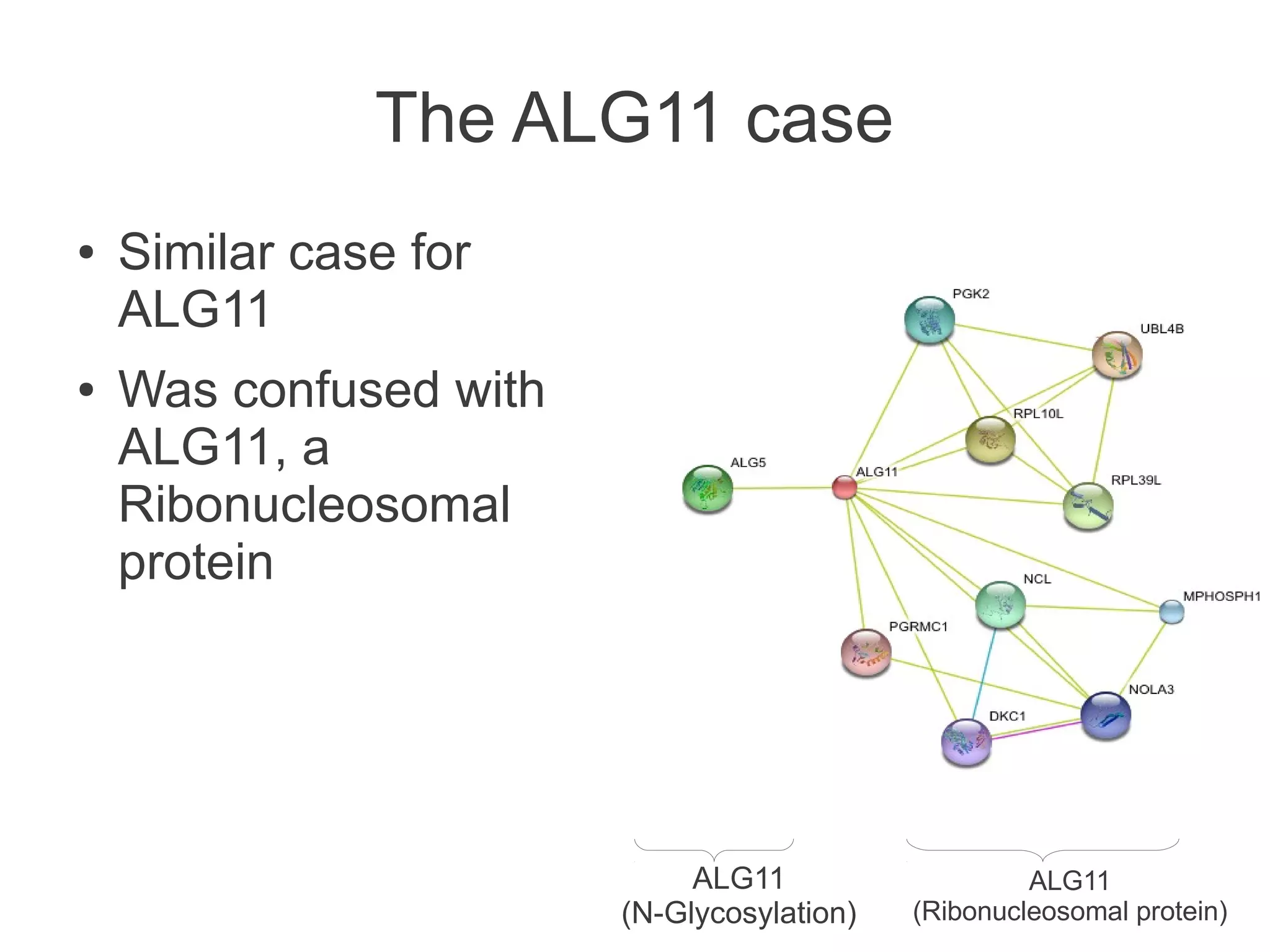
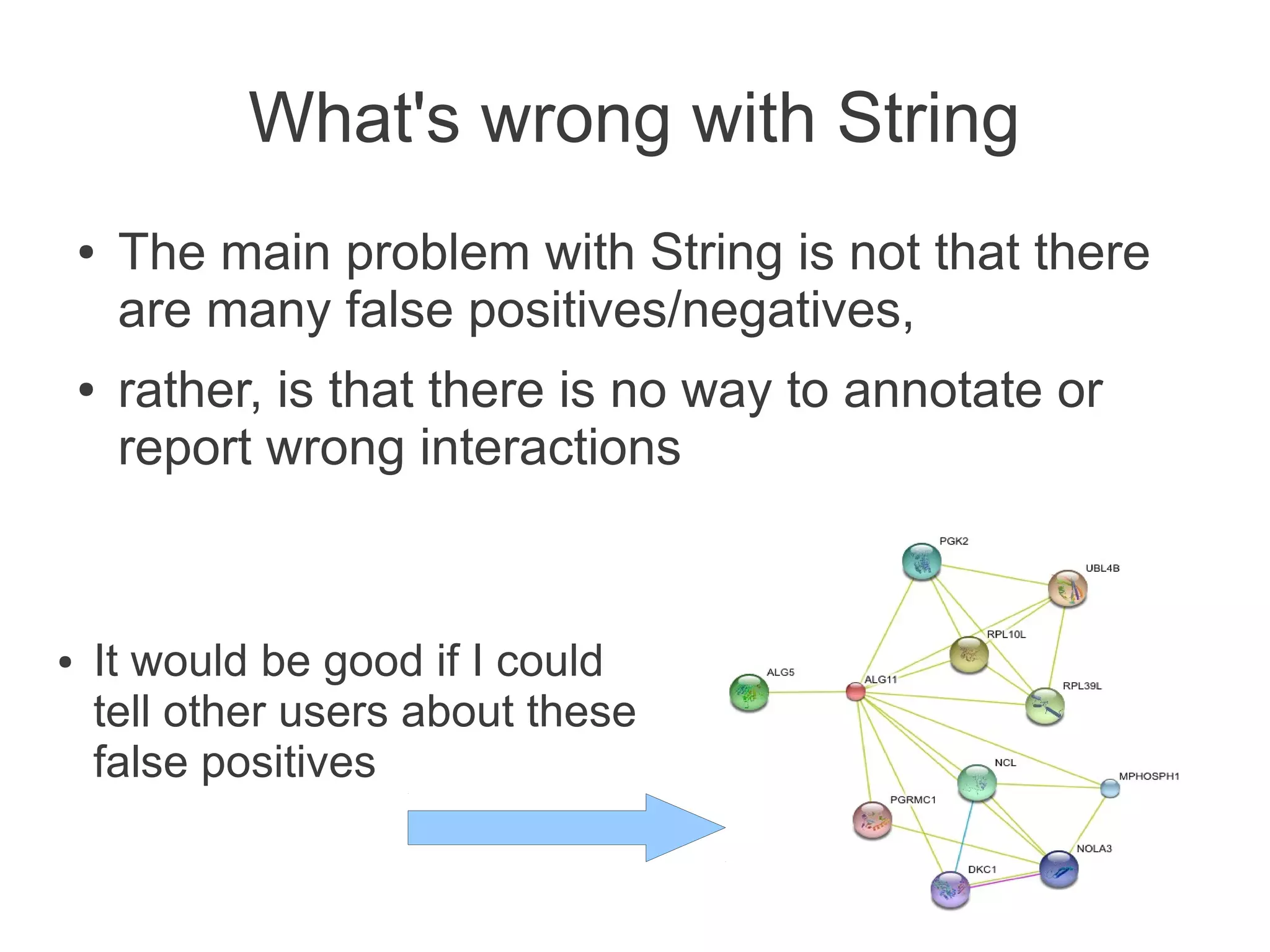
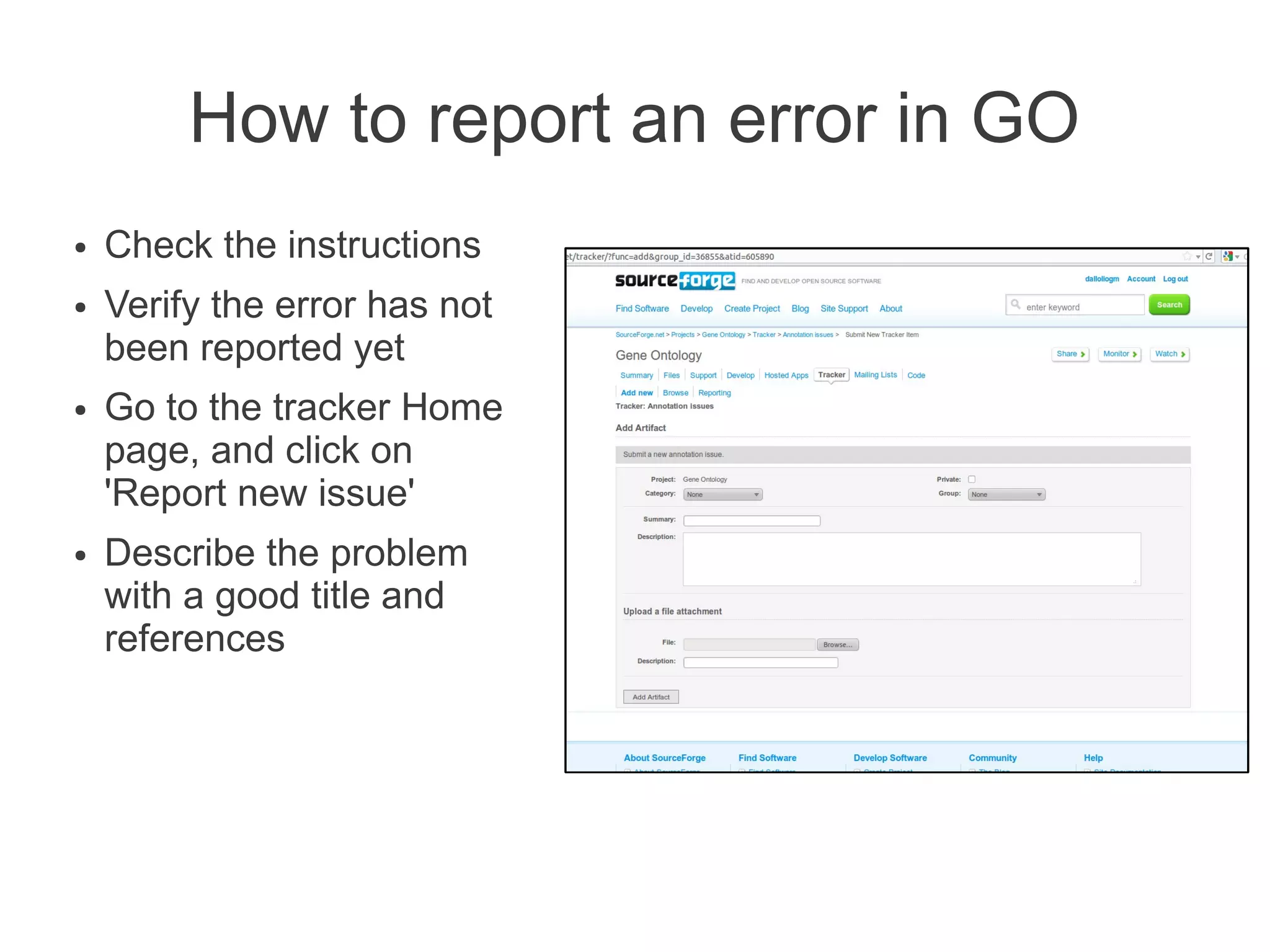
The document summarizes a talk given about annotating the N-glycosylation pathway in scientific databases. The speaker annotated the pathway in Reactome by providing details for over 100 reactions from literature. This revealed errors in other databases and variable interpretations of terms. Reporting issues to public trackers can help fix errors. Database annotations are imperfect and interpreting pathways requires considering evidence sources.