Embed presentation
Downloaded 55 times











![Summary Increased use of ontologies is inevitable as data volumes grow. UniProt has (or is in the process of introducing) several ontologies. What data will be "ontologized" and how detailed the ontologies are depends on your feedback ! [email_address]](https://image.slidesharecdn.com/uniprot-ontologies-18420/85/UniProt-Ontologies-24-320.jpg)

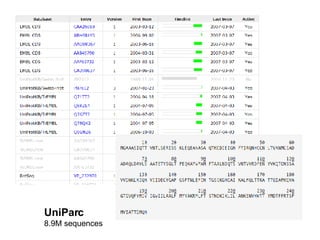
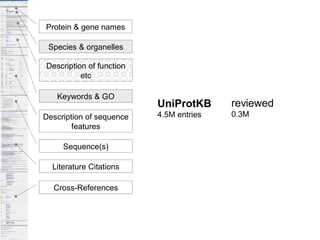
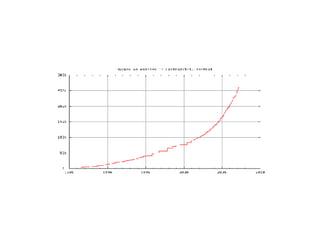
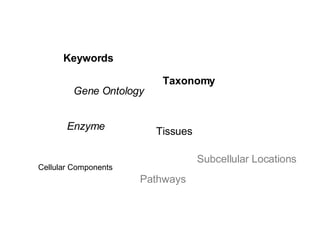
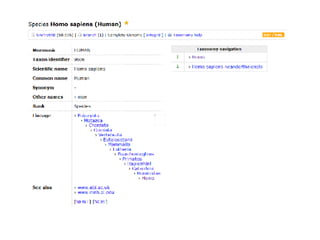
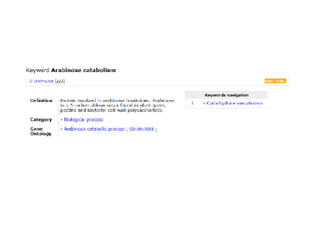
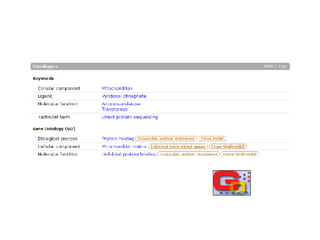
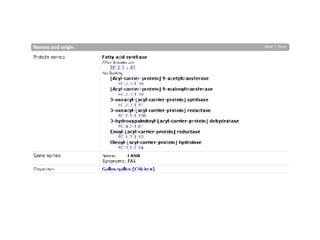
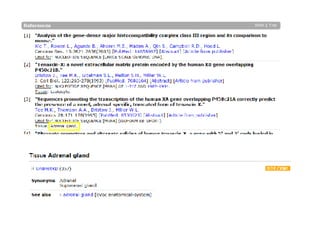
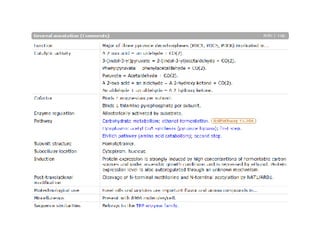
UniProt is a large life sciences database that uses several ontologies like the Gene Ontology, keywords, taxonomy, and pathways to provide consistent navigation and aggregation of its data on protein sequences, functions, and features. The document discusses how ontologies help organize UniProt's large amount of data and provide benefits like auto-completion, set-oriented views, and potential for automation. It also seeks feedback on which additional data from UniProt could be organized through more detailed ontologies.























![Summary Increased use of ontologies is inevitable as data volumes grow. UniProt has (or is in the process of introducing) several ontologies. What data will be "ontologized" and how detailed the ontologies are depends on your feedback ! [email_address]](https://image.slidesharecdn.com/uniprot-ontologies-18420/85/UniProt-Ontologies-24-320.jpg)
