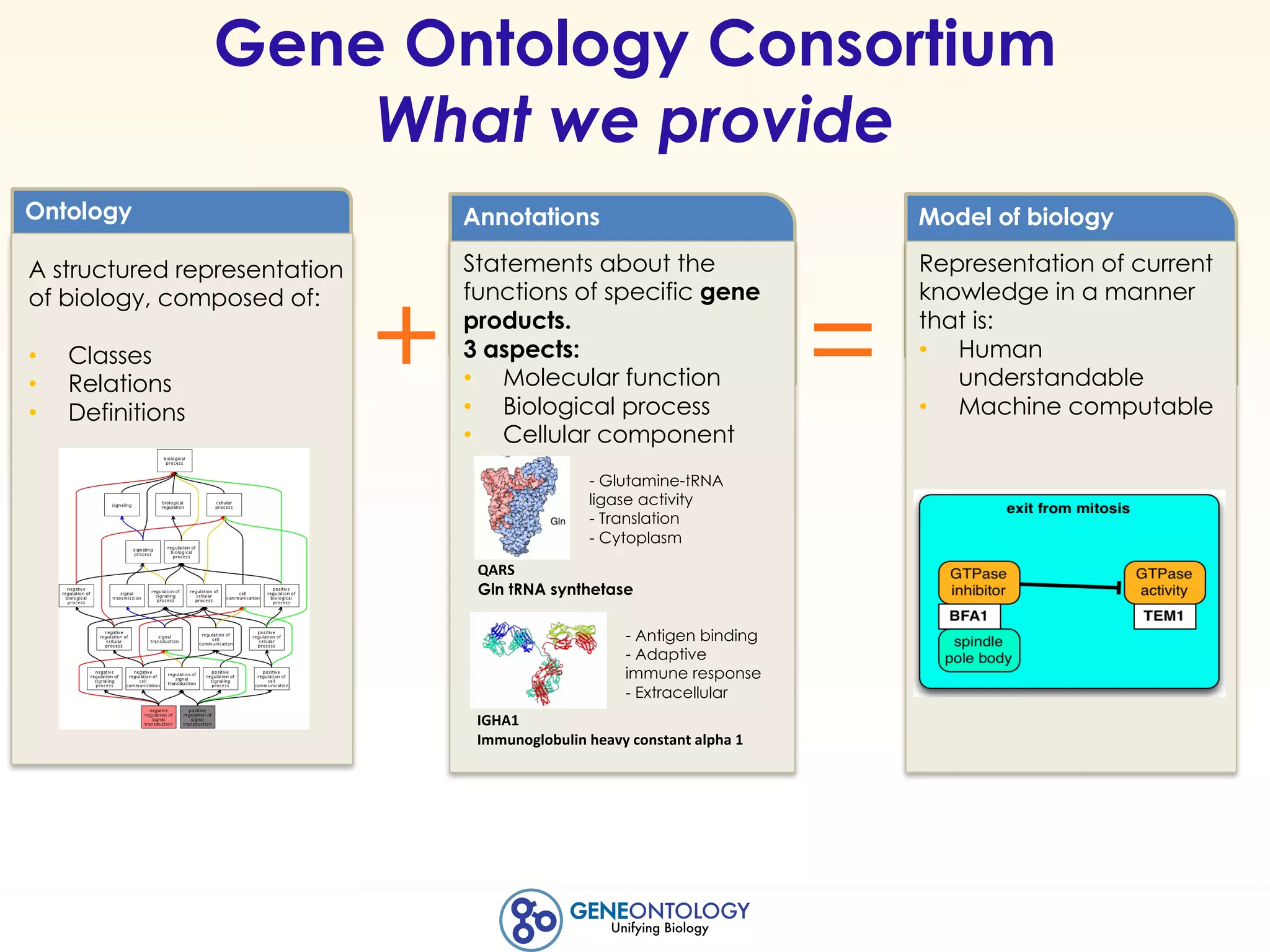
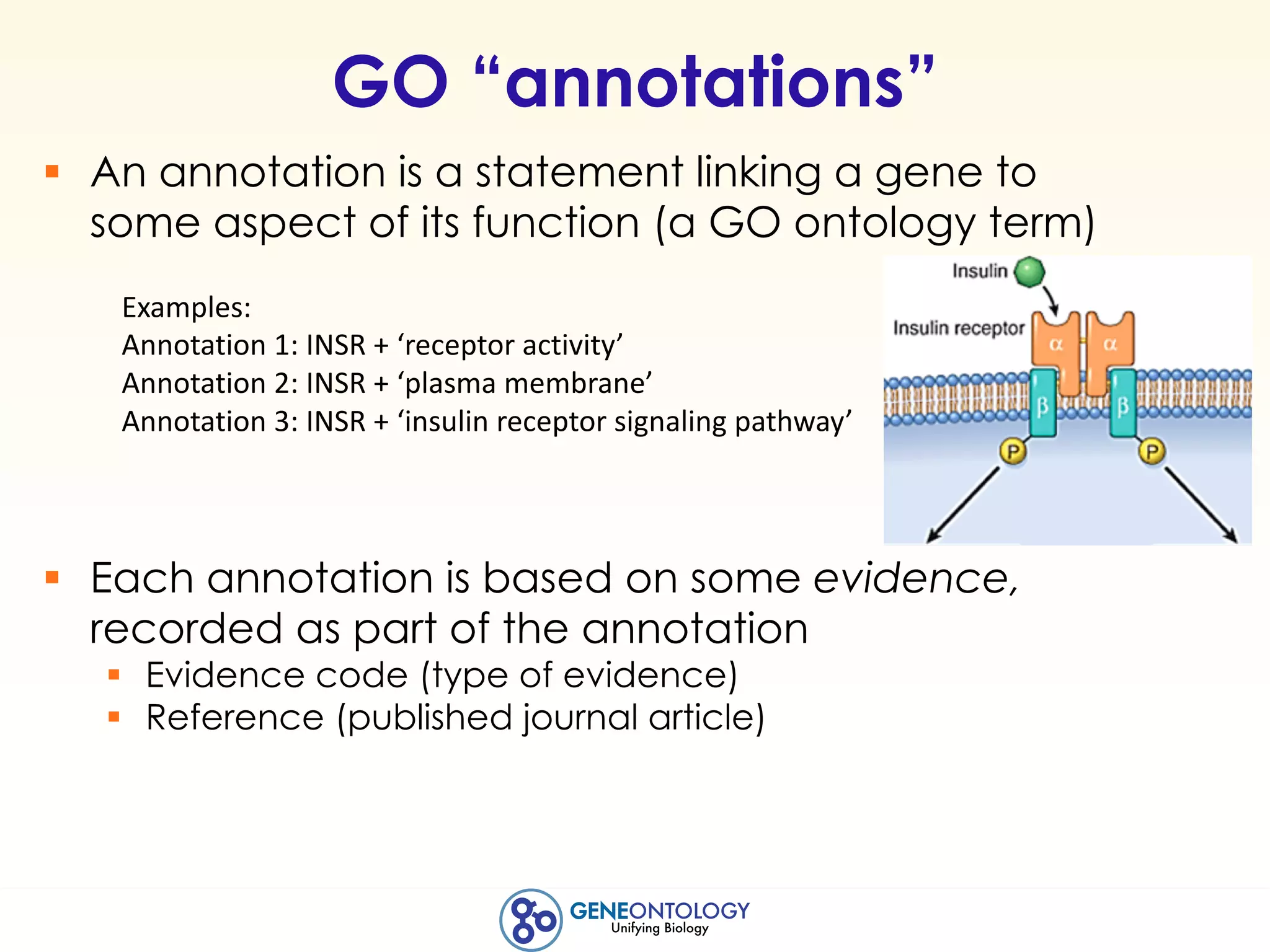
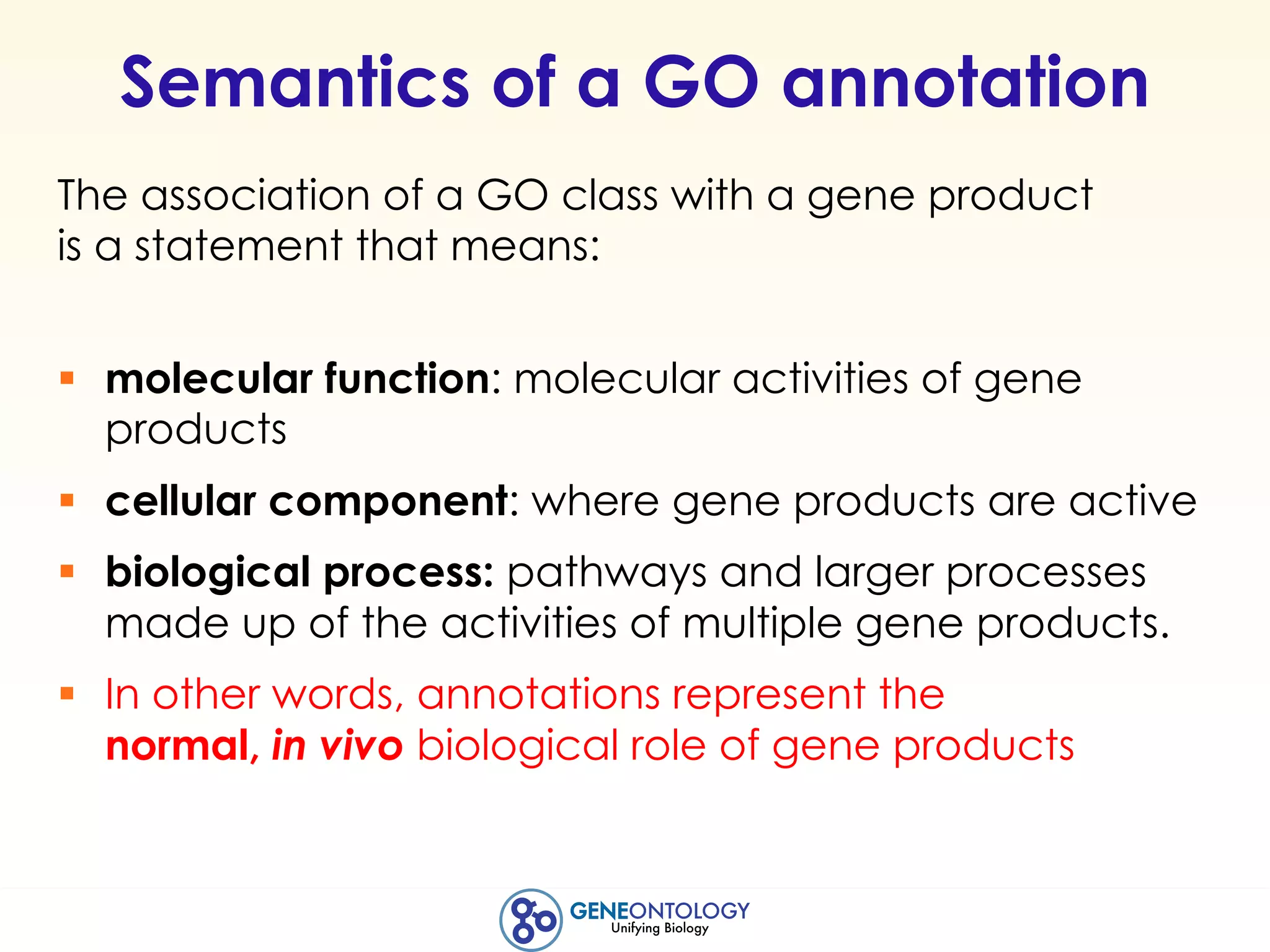
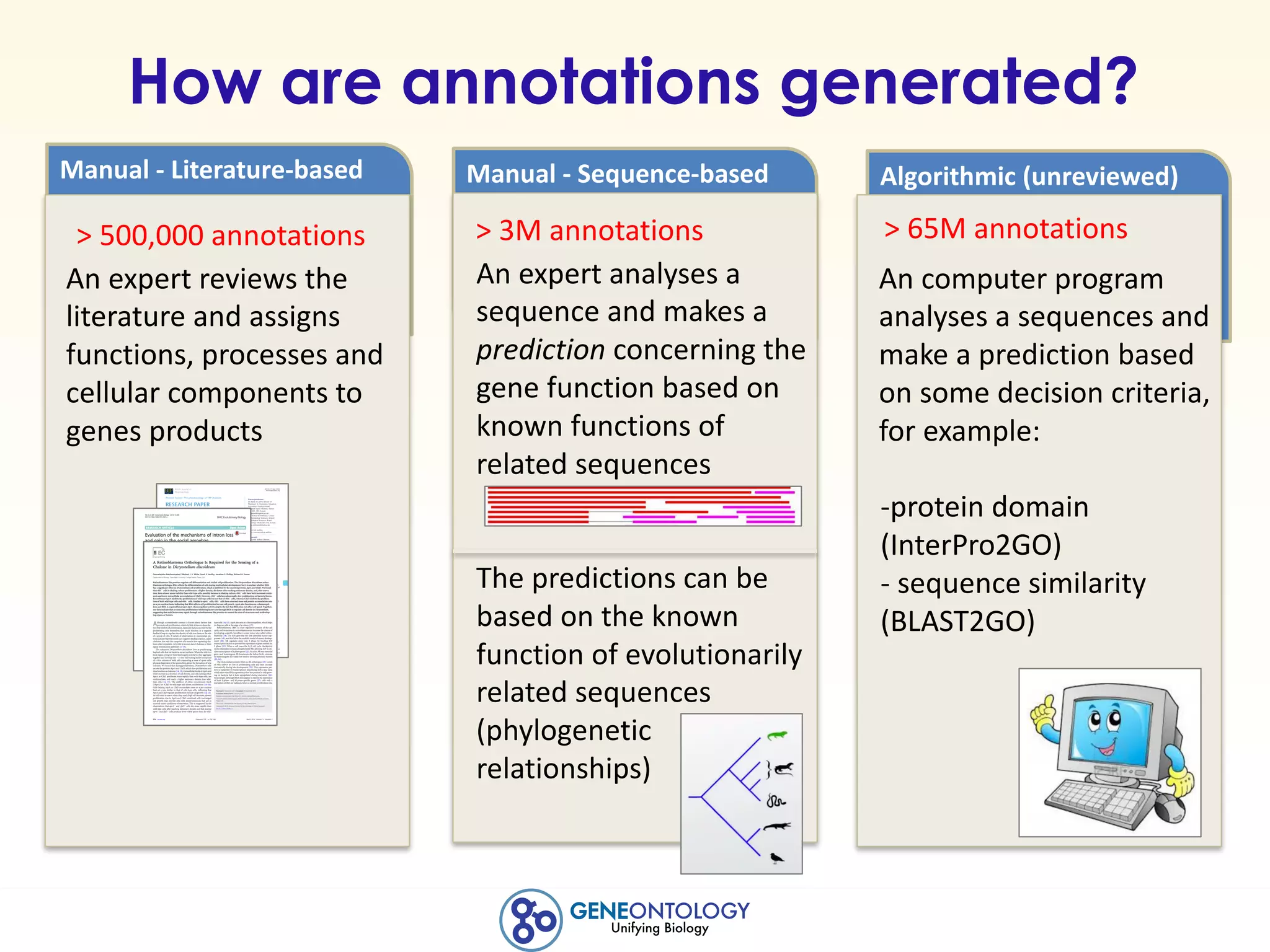
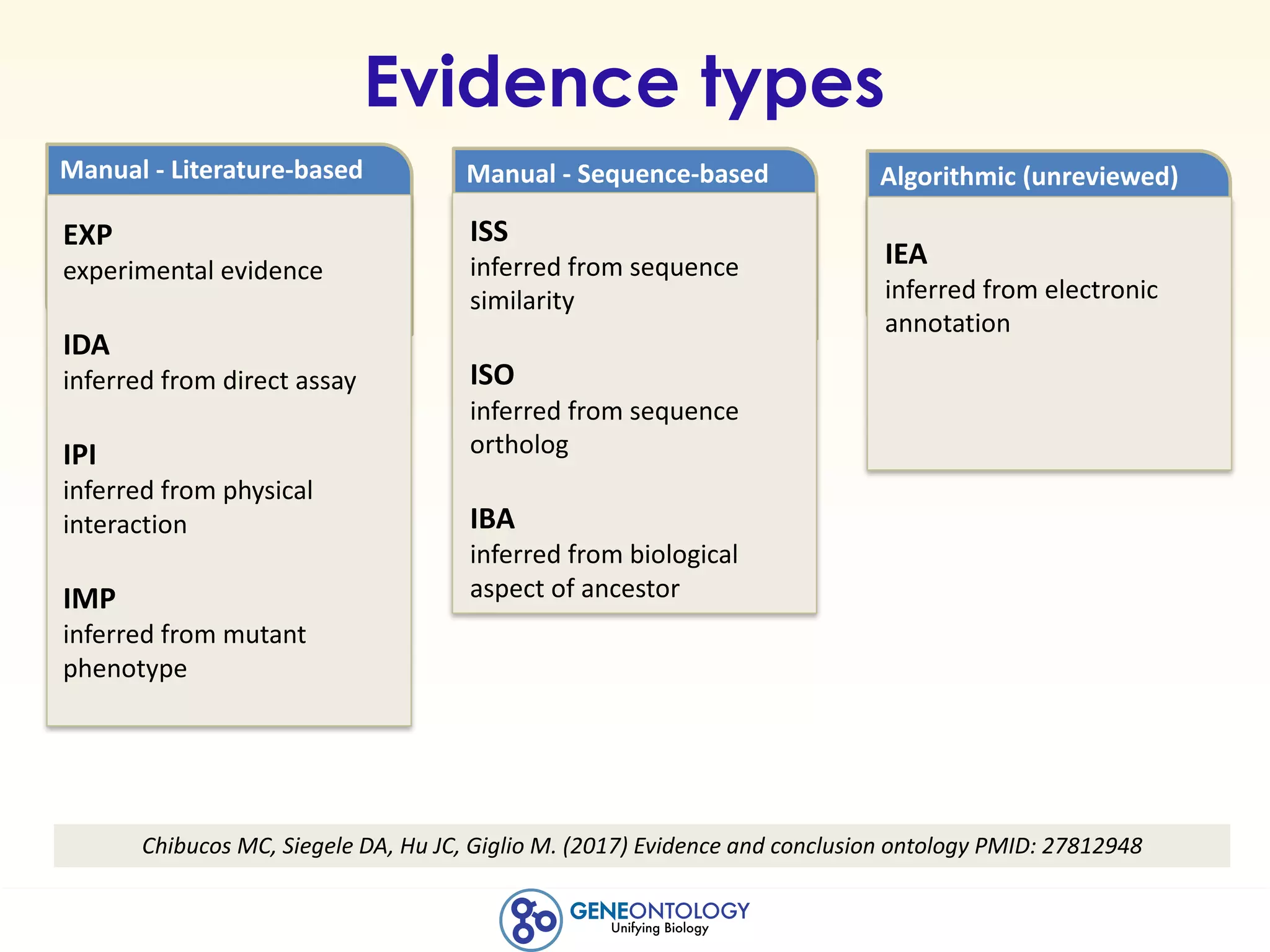
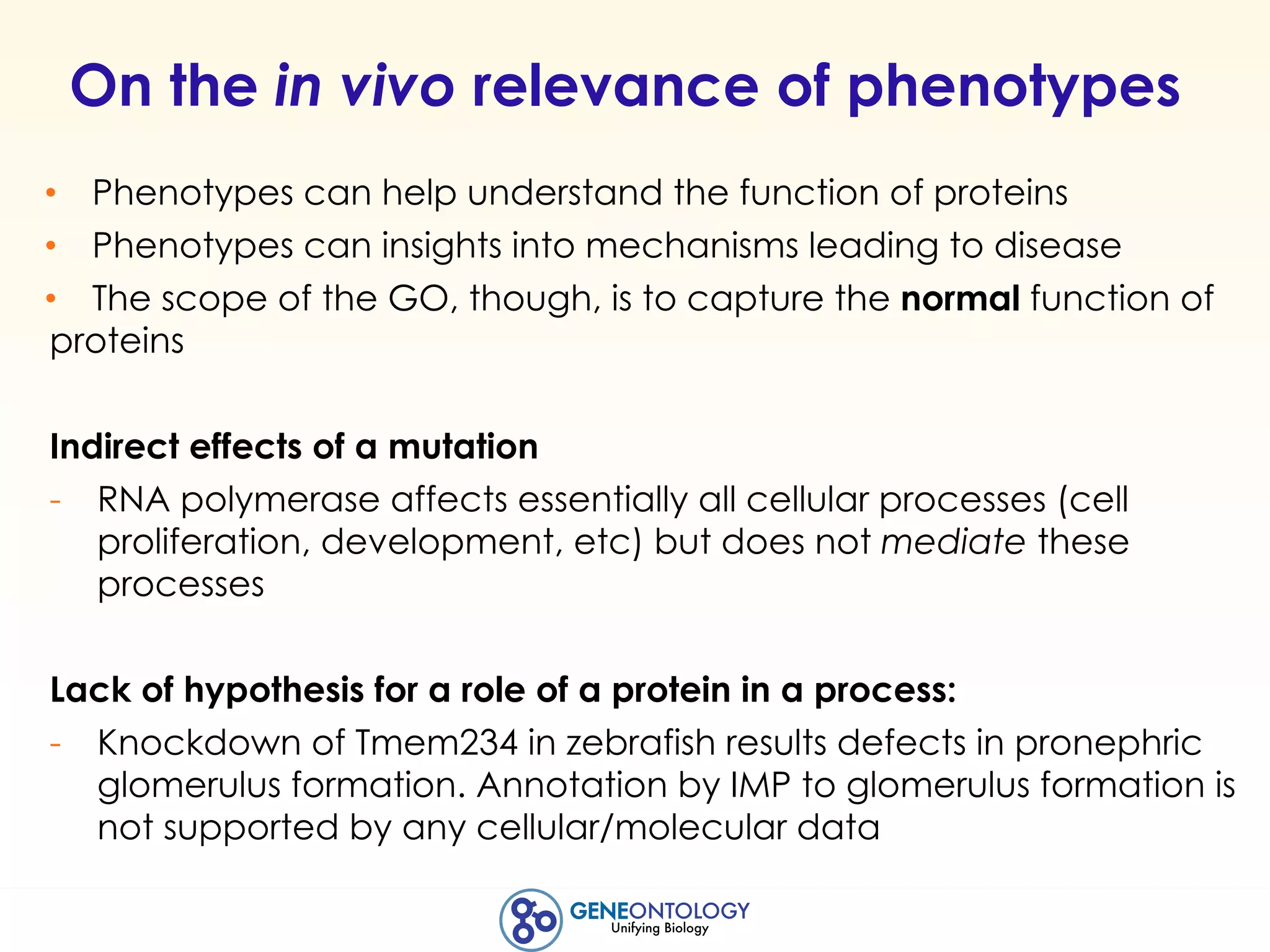
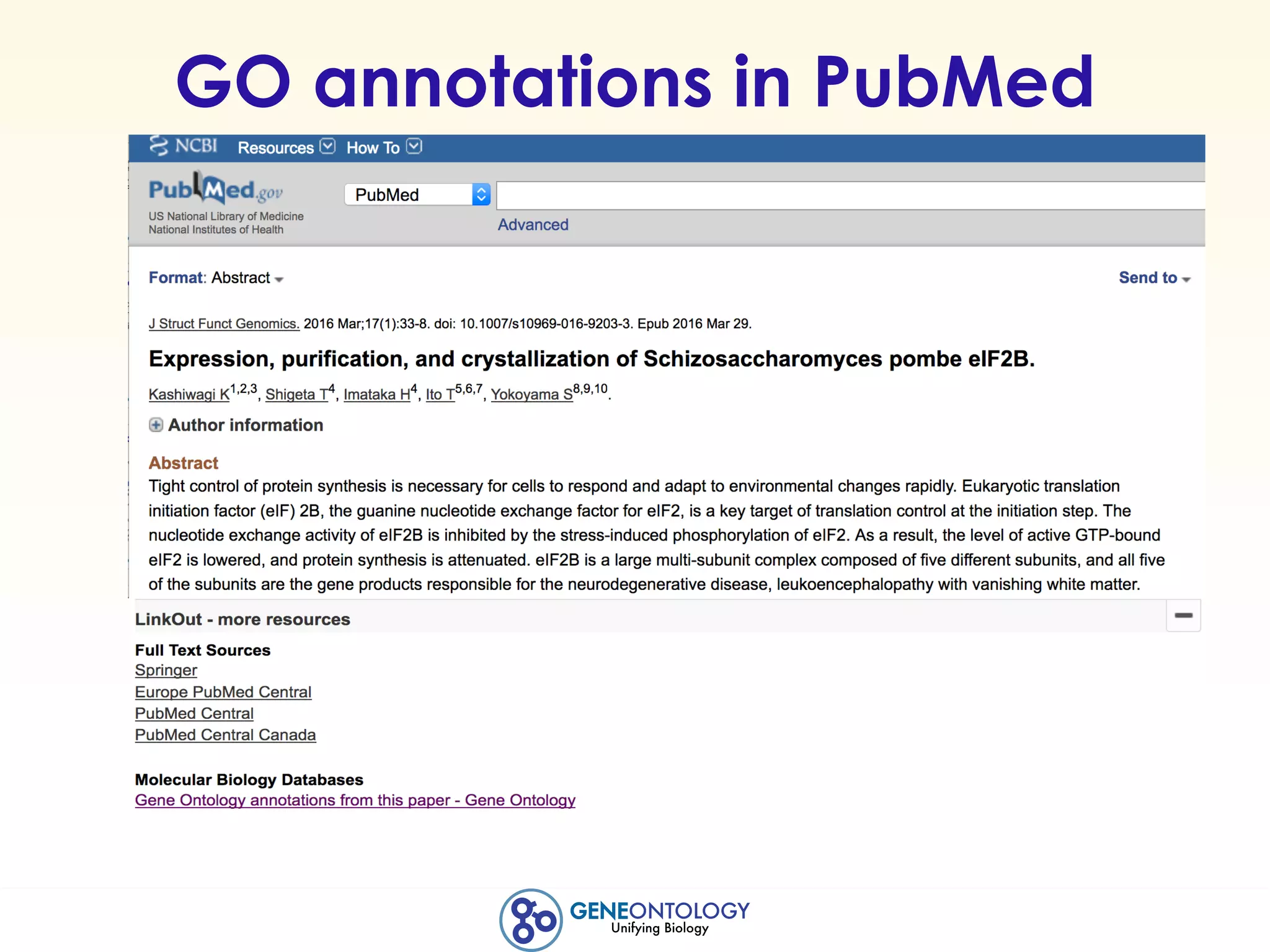
This document discusses Gene Ontology (GO) annotations, which are statements linking genes to aspects of their functions as represented by GO terms. Annotations are generated through manual literature curation, manual sequence analysis, or algorithmic prediction. They are assigned evidence codes and references and represent the normal biological roles of gene products based on molecular function, biological process, and cellular component. Guidelines are provided for producing high-quality, literature-based annotations that accurately reflect experimental conclusions and biological context.