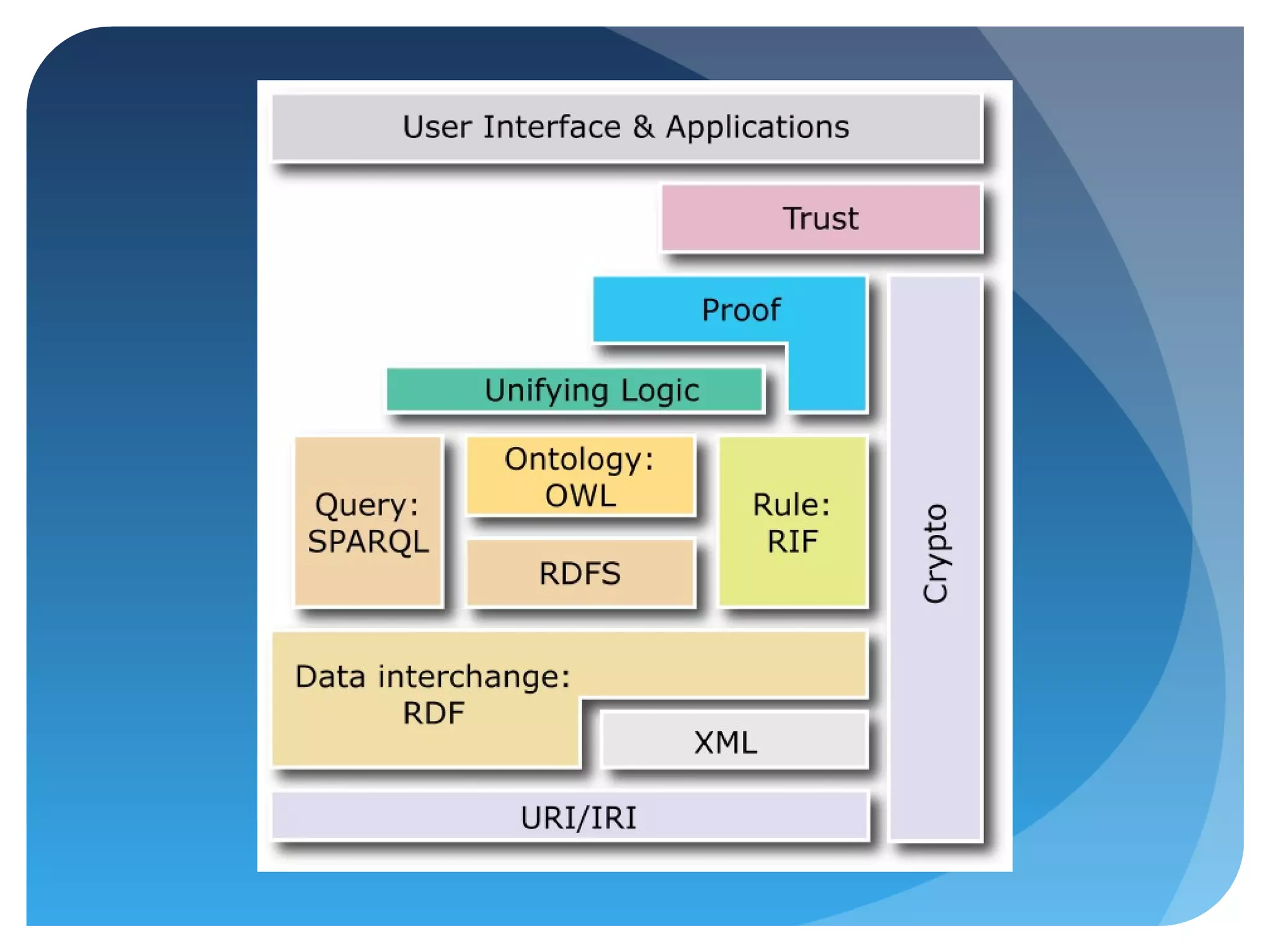
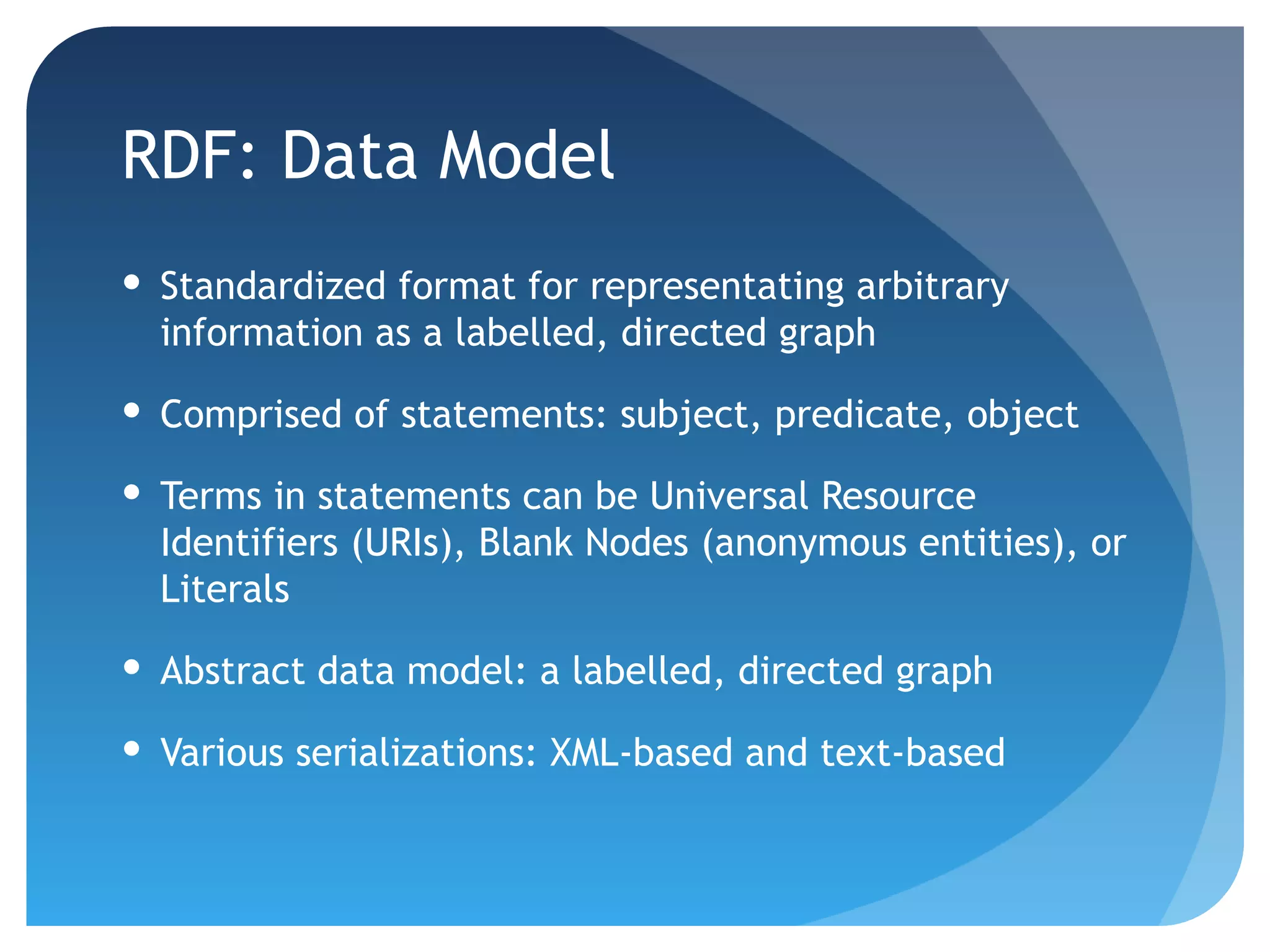
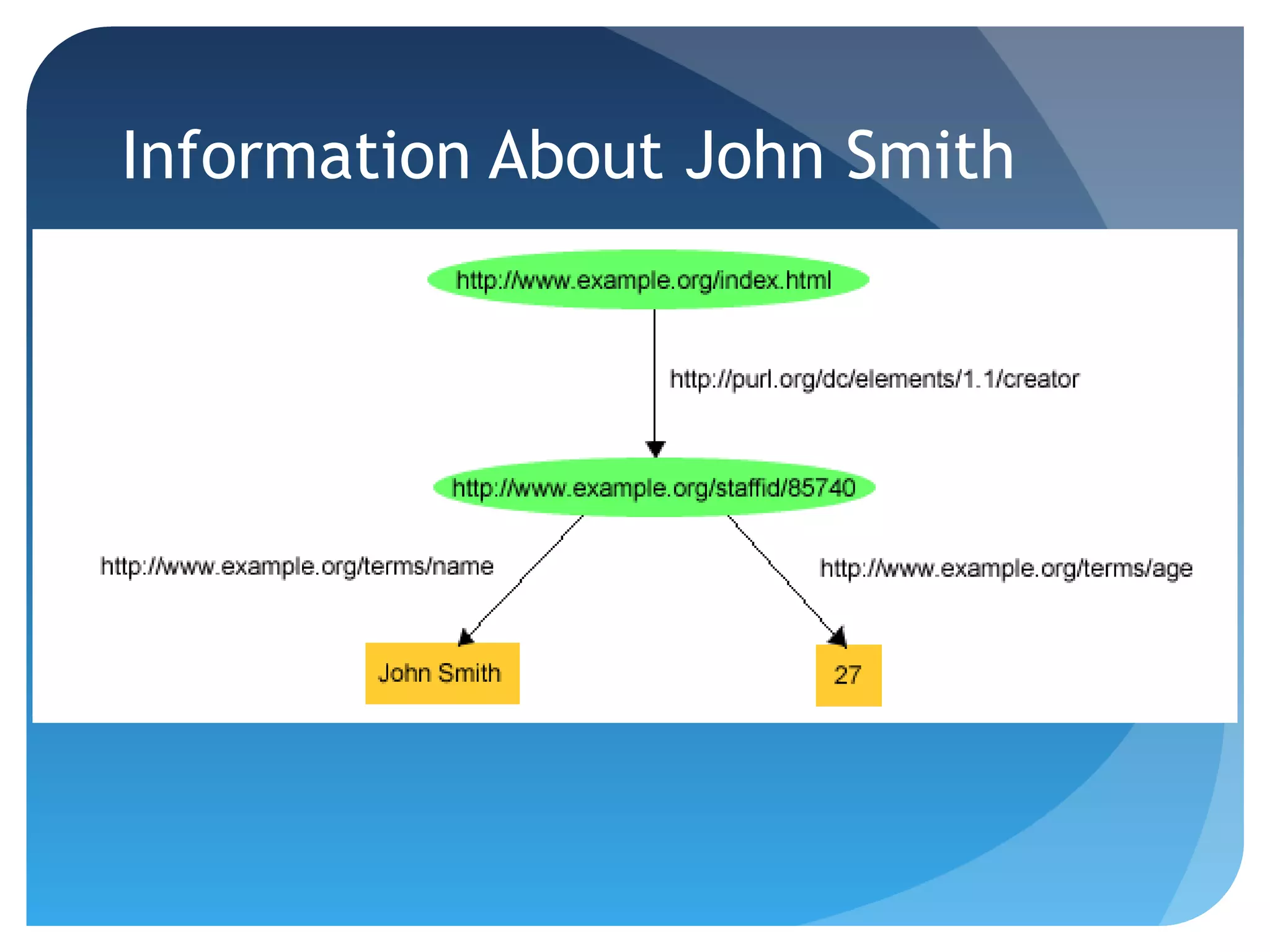
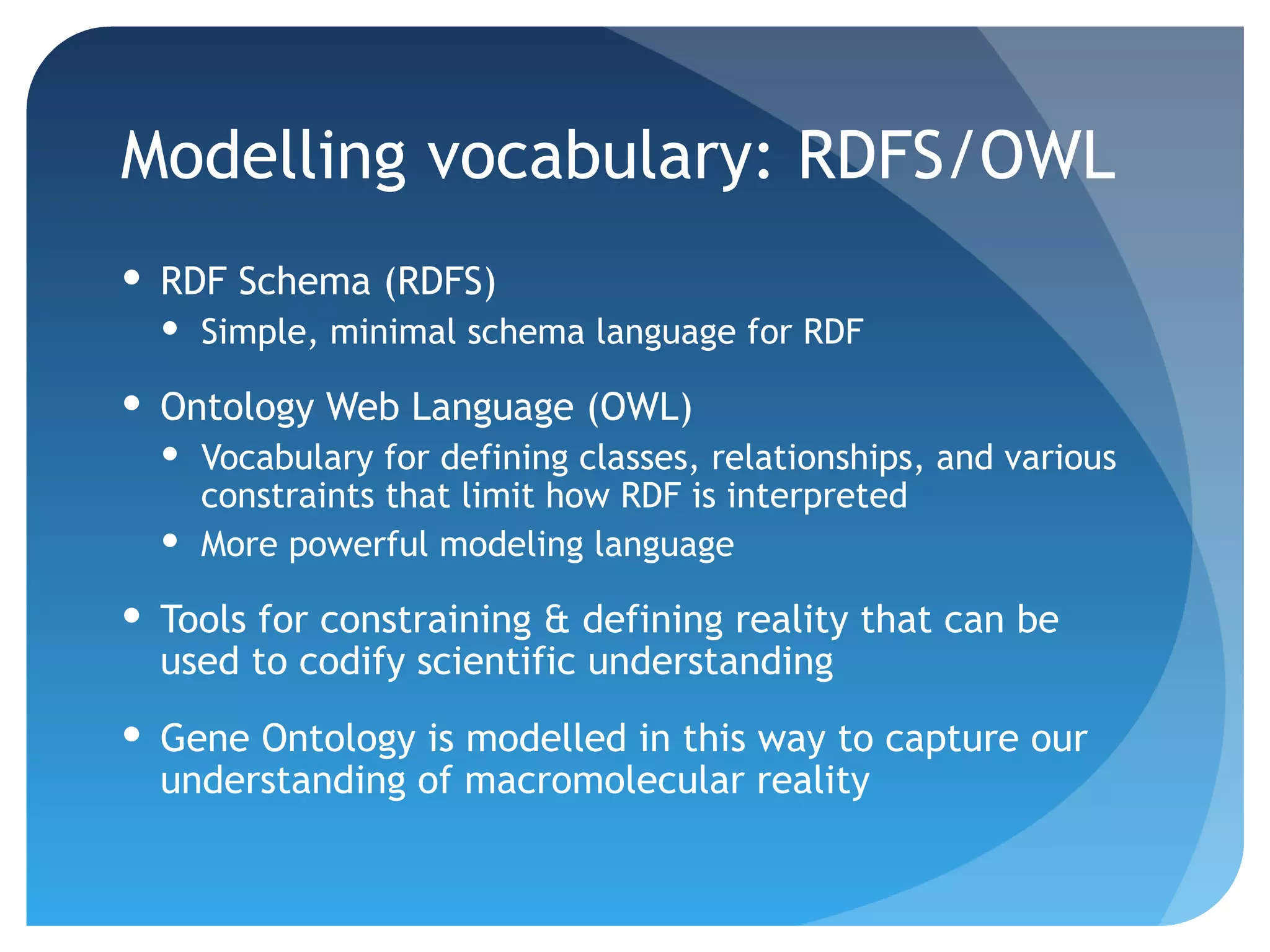
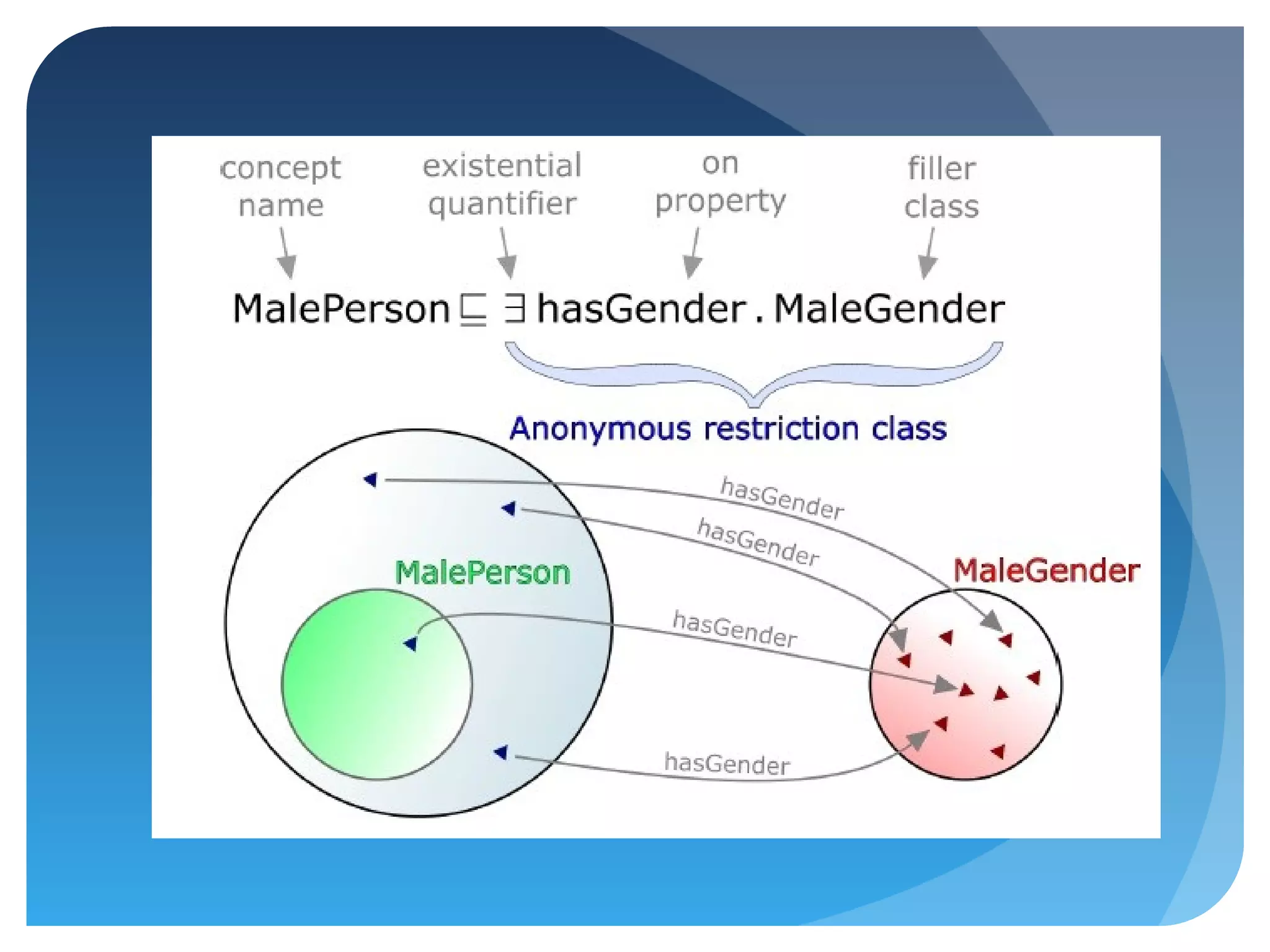
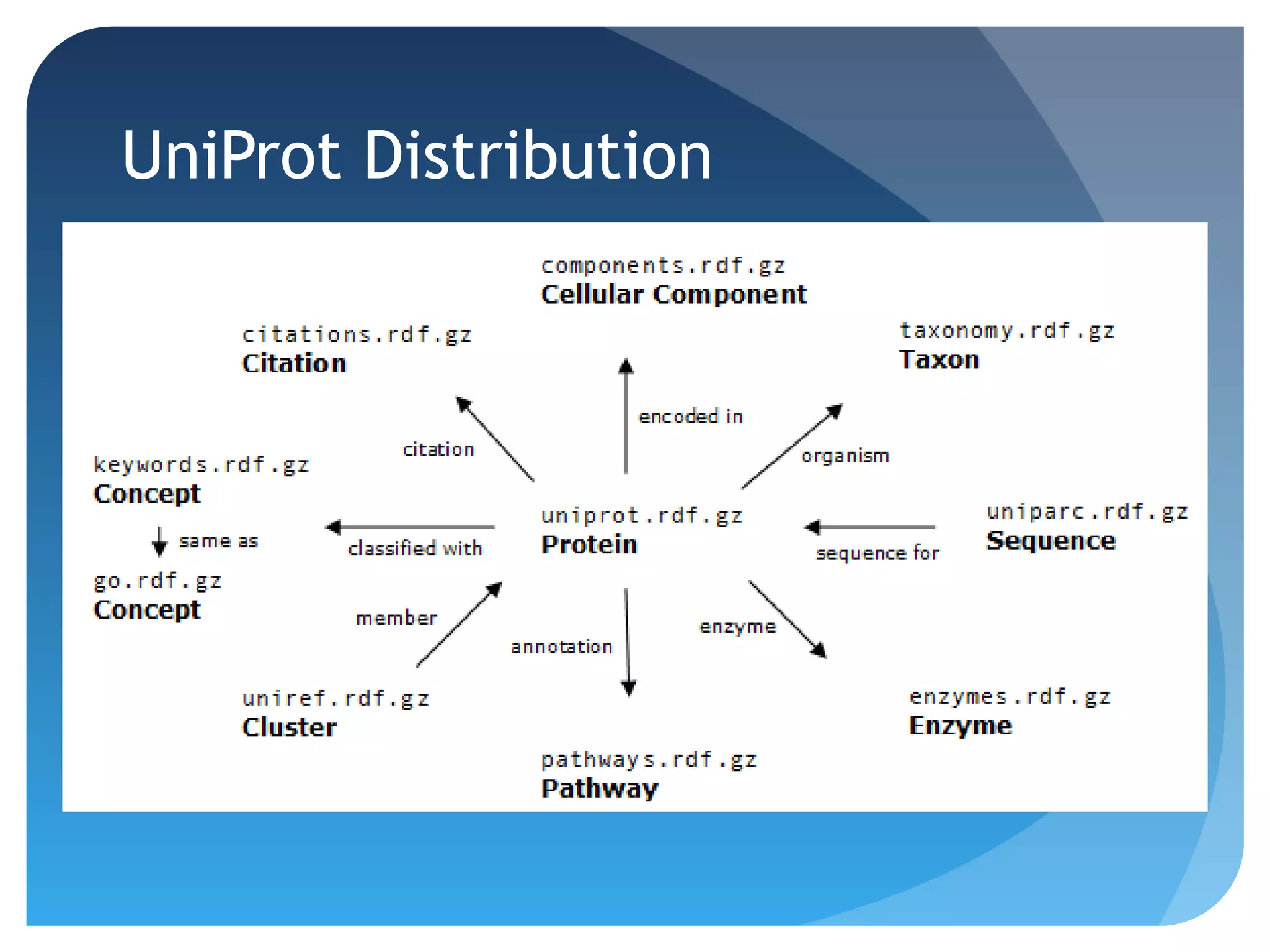
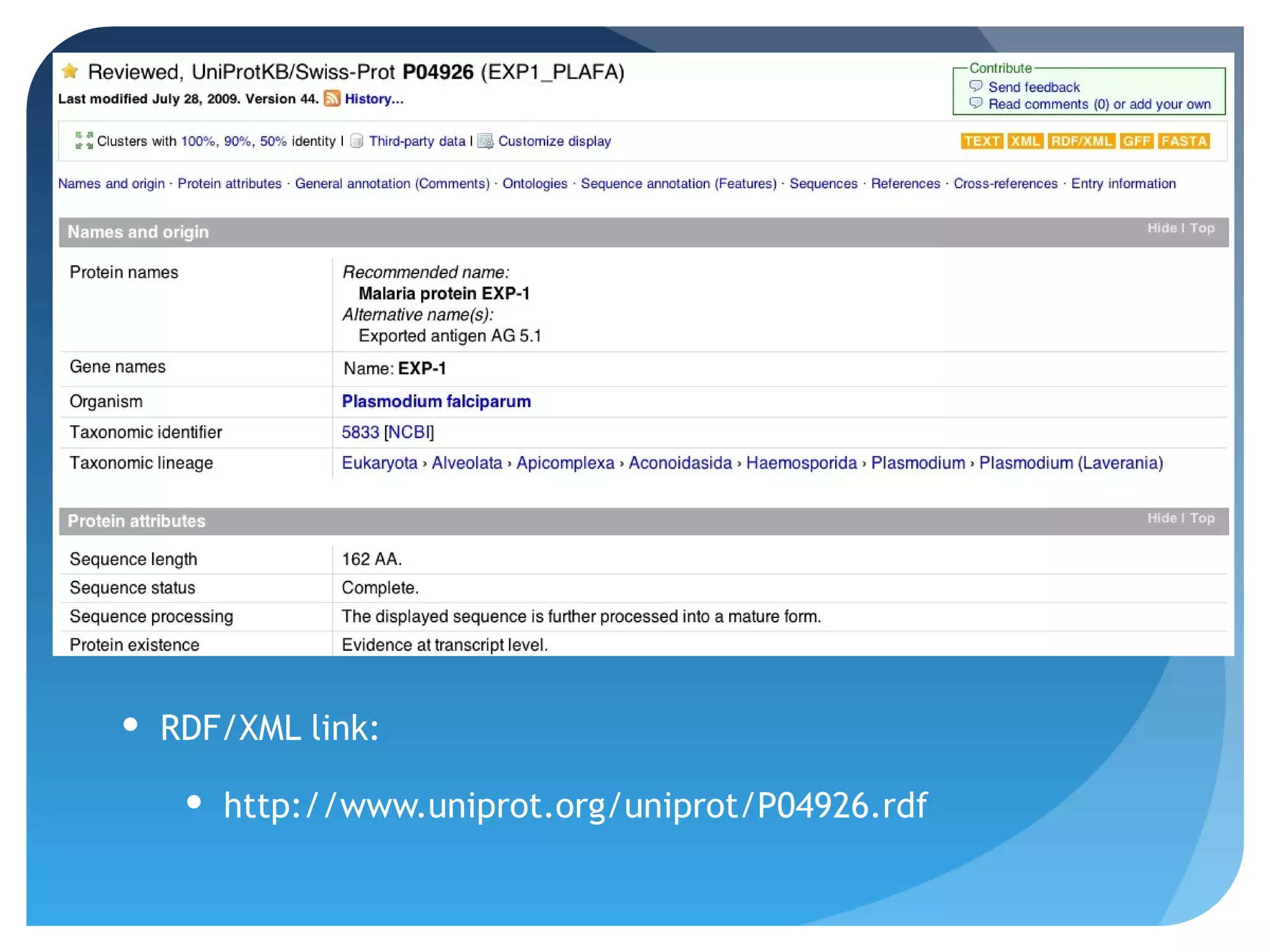
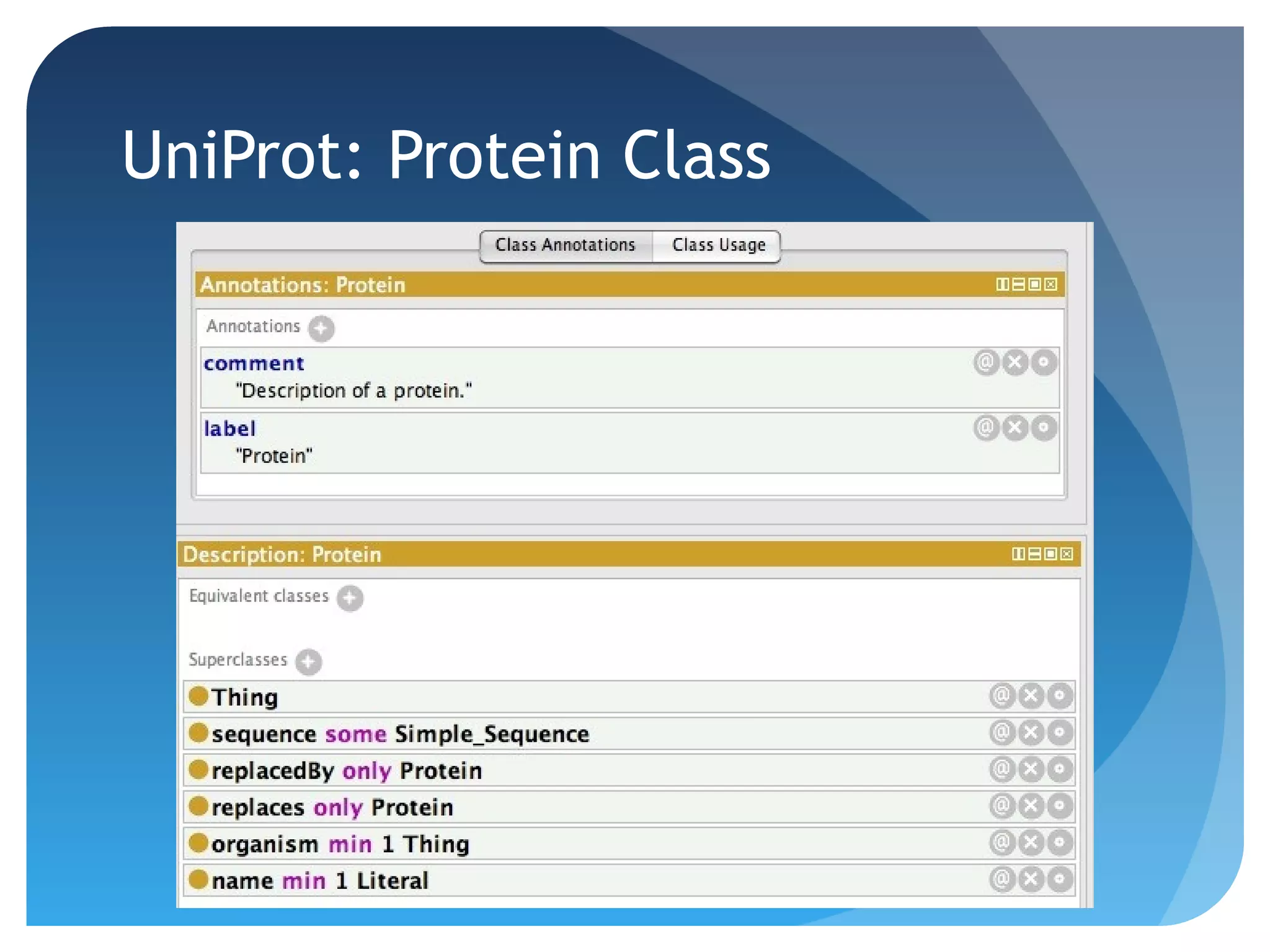
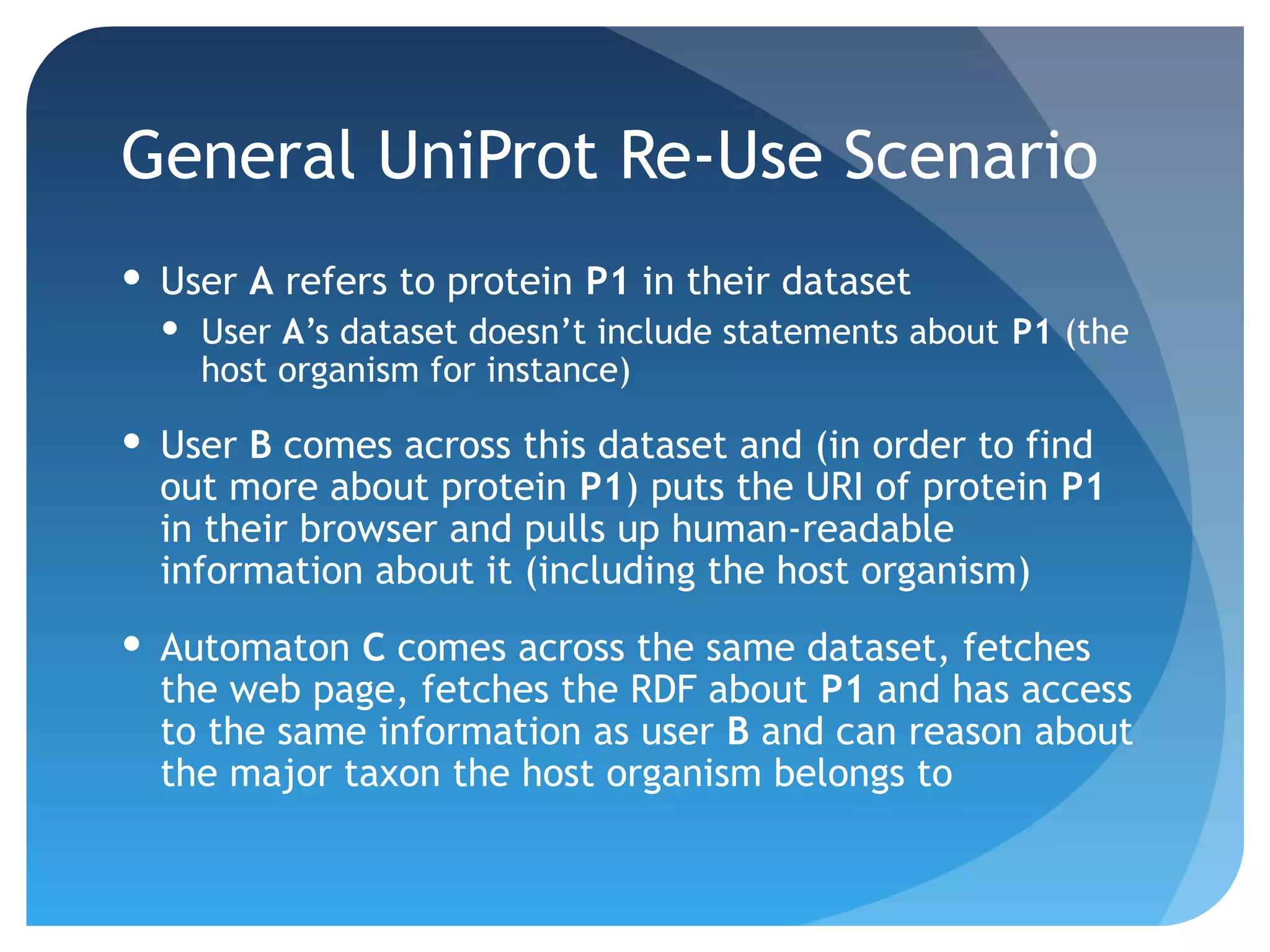
This document discusses how UniProt addresses challenges of integrating protein data from various sources by adopting Semantic Web technologies like RDF, OWL, and SPARQL. It describes how UniProt data is modeled and distributed as RDF to allow for automated querying and integration with external data sources. By using PURLs and RDF, UniProt aims to provide a comprehensive, standardized representation of protein data that facilitates data sharing and reuse across scientific research.