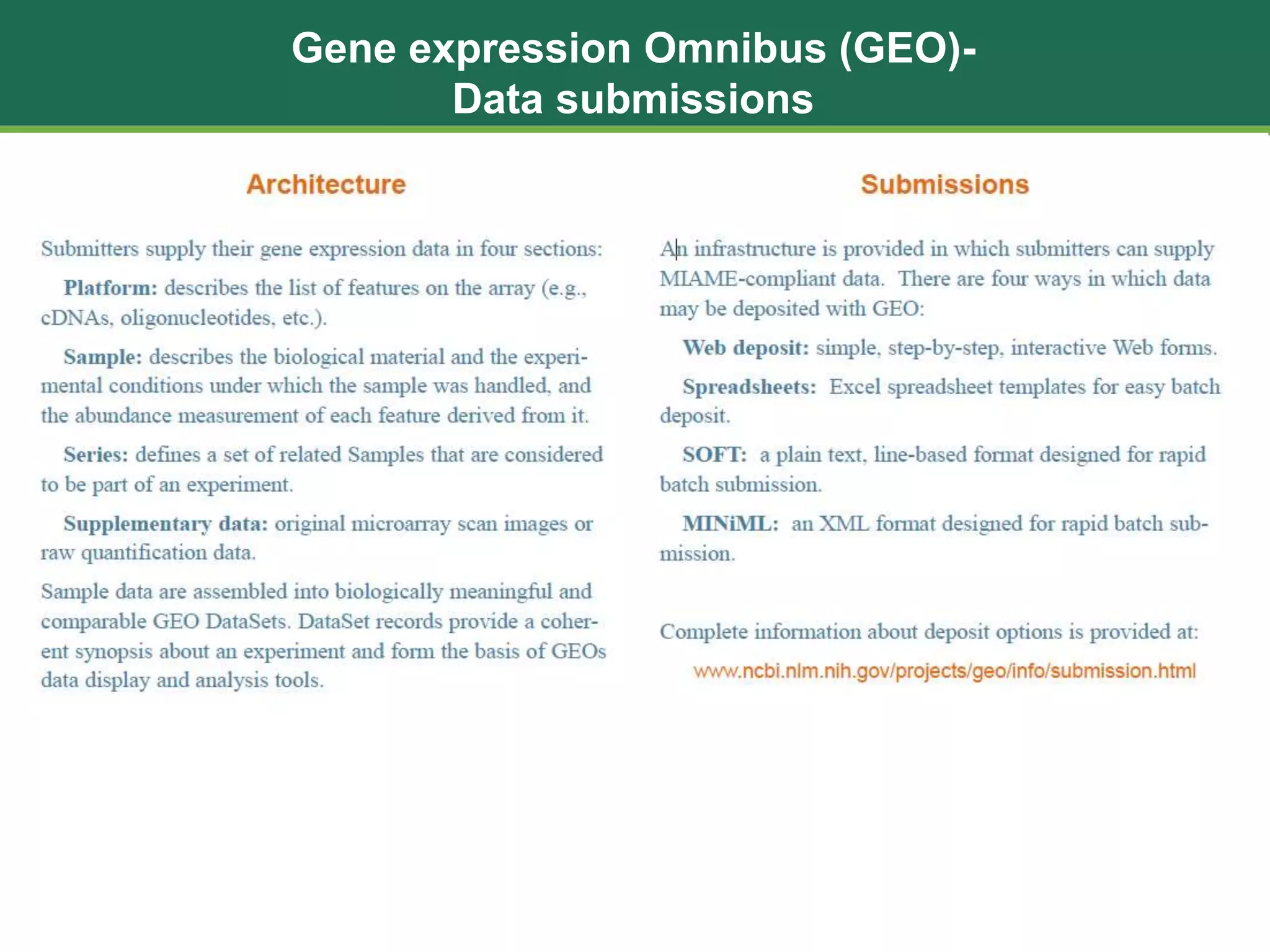
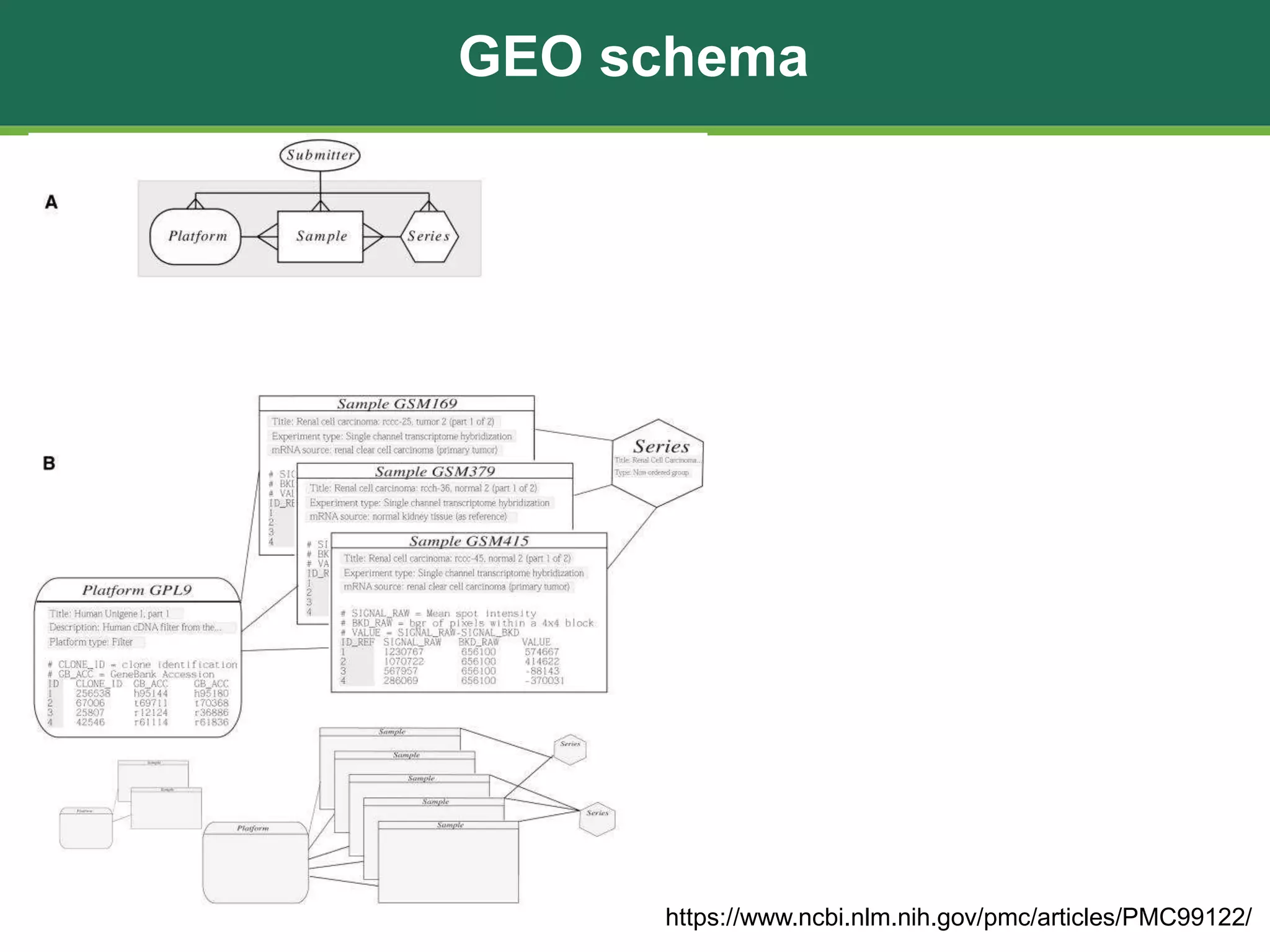
The document discusses the Gene Expression Omnibus (GEO) database, which is a public repository maintained by the National Center for Biotechnology Information that archives and freely distributes gene expression and other high-throughput functional genomics data submitted by the research community. It outlines the background and purpose of GEO, describes the types of data it contains including platforms, samples, and series. It also lists 10 ways that researchers can utilize and analyze data from GEO, such as performing searches, downloading data files, and using online analysis tools to visualize and compare gene expression data.