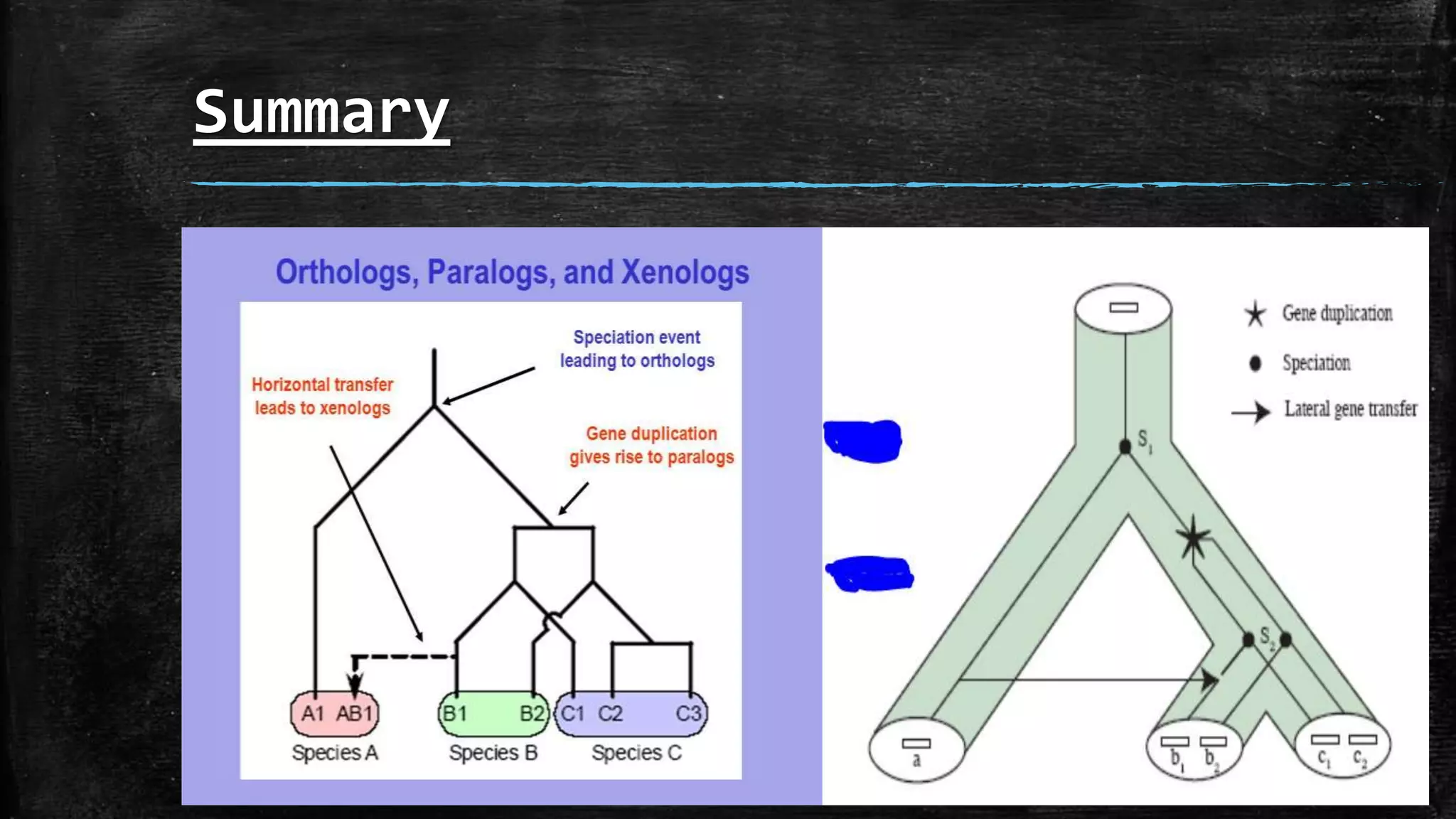
Orthologs are homologous sequences that descended from a common ancestral sequence after a speciation event separated two species. Paralogs are homologous sequences related through a gene duplication event in a common ancestor. Xenologs are homologous sequences resulting from horizontal gene transfer between two organisms. The document discusses these three types of sequence homology - orthologs, paralogs, and xenologs - which arise from different evolutionary events involving speciation, duplication, and horizontal transfer of genes.