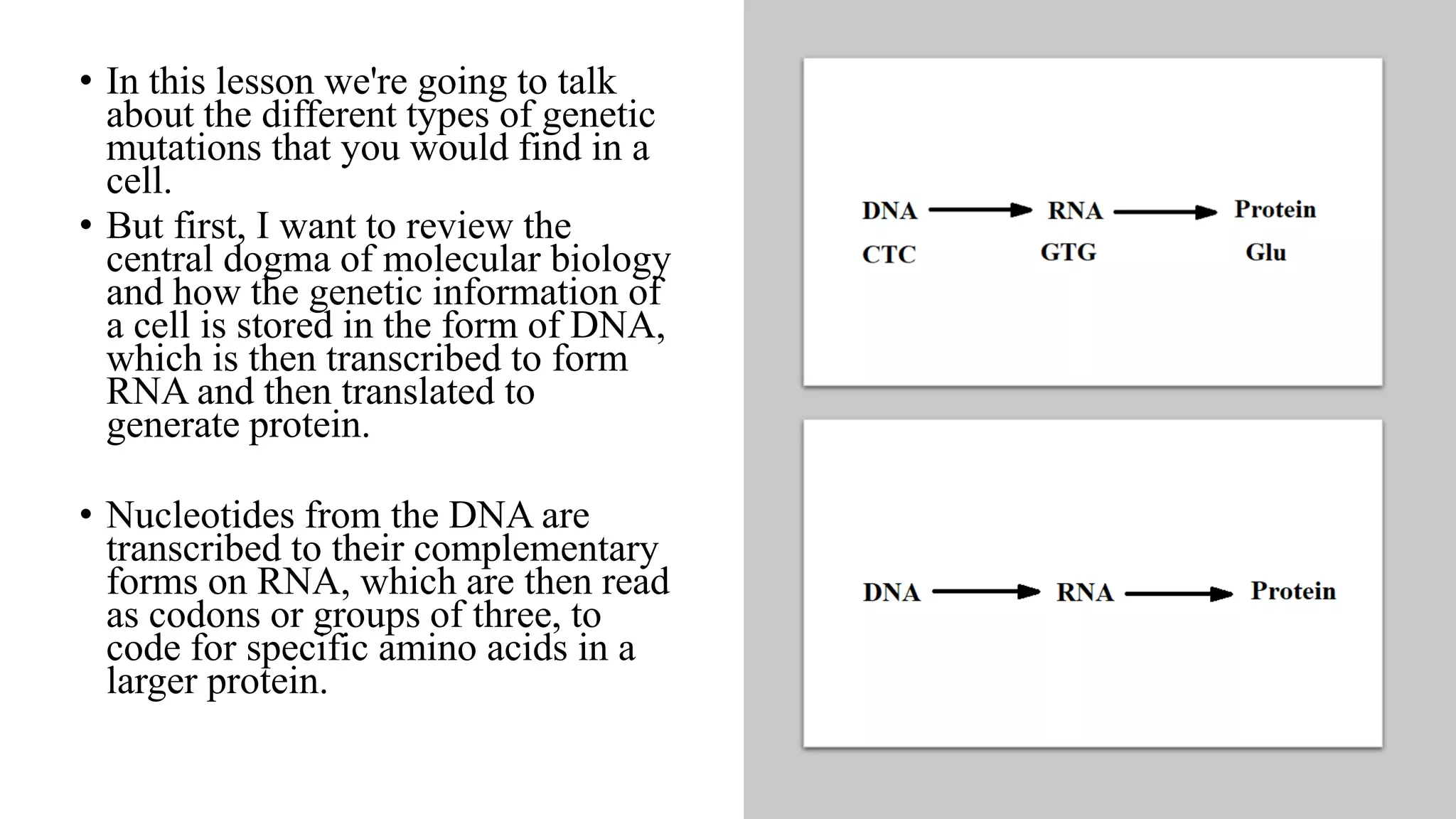
1. The document discusses different types of genetic mutations including point mutations, frameshift mutations, nonsense mutations, missense mutations, silent mutations, conservative mutations, and nonconservative mutations.
2. Mutations originate at the DNA level but their effects are seen at the protein level. Mutations can be classified based on their effects on DNA or proteins.
3. The main types of mutations are point mutations and frameshift mutations at the DNA level, and missense and nonsense mutations at the protein level.