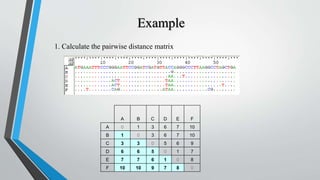
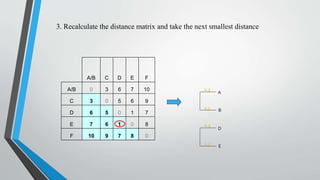
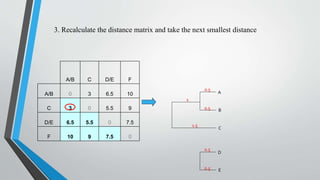
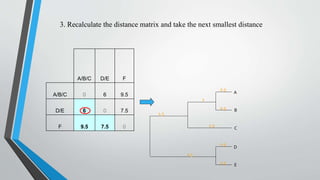
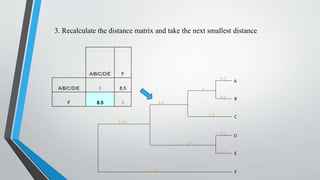
UPGMA is an algorithm for constructing phylogenetic trees from distance matrix data. It works by sequentially clustering the two closest groups at each step, computing distances between new clusters and other groups as the average of all pairwise distances. UPGMA assumes a molecular clock and produces rooted, ultrametric trees reflecting phenotypic similarities rather than true evolutionary relationships.









![Applications
• In ecology, it is one of the most popular methods for the classification of sampling units (such
as vegetation plots) on the basis of their pairwise
similarities in relevant descriptor variables (such as species composition).[3]
• In bioinformatics, UPGMA is used for the creation of phenetic trees (phenograms). UPGMA
was initially designed for use in protein
electrophoresis studies, but is currently most often used to produce guide trees for more sophi
sticated algorithms. This algorithm is for example
used in sequence alignment procedures, as it proposes one order in which the sequences will
be aligned. Indeed, the guide tree aims at grouping
the most similar sequences, regardless of their evolutionary rate or phylogenetic affinities, an
d that is exactly the goal of UPGMA.[4]
• In phylogenetics, UPGMA assumes a constant rate of evolution (molecular clock hypothesis),
and is not a wellregarded method for inferring
relationships unless this assumption has been tested and justified for the data set being used.](https://image.slidesharecdn.com/upmafinal-170419184035/85/UPGMA-10-320.jpg)