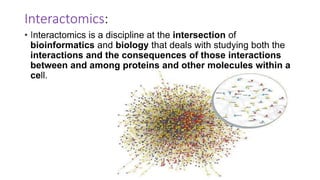
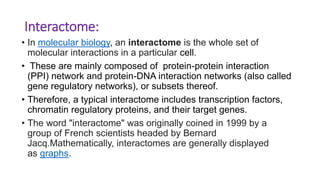
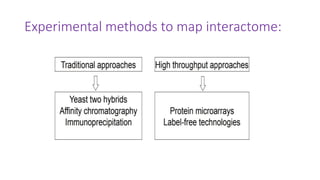
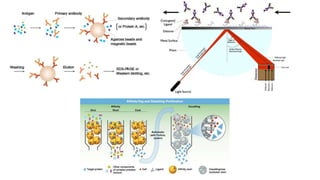
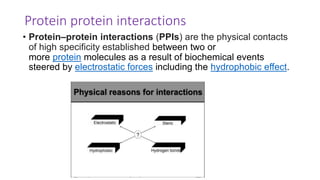
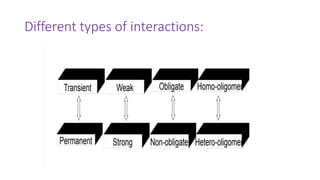
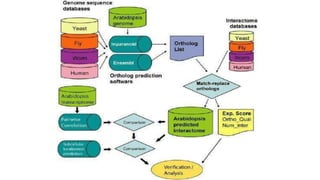
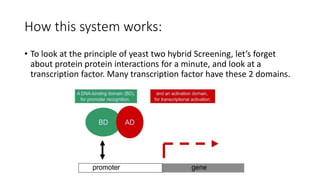
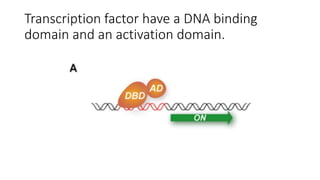
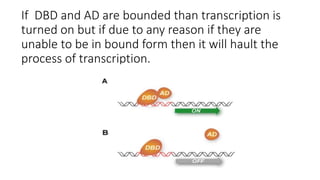
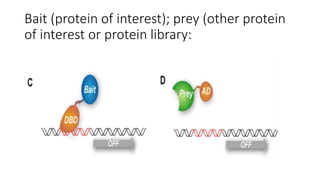
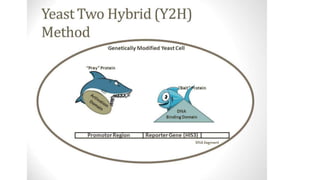
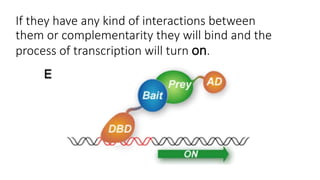
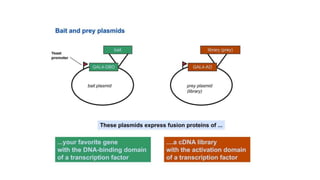
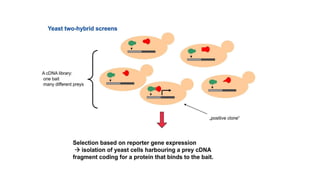
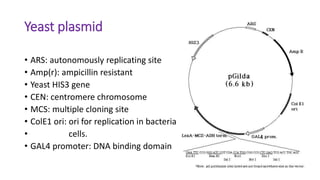
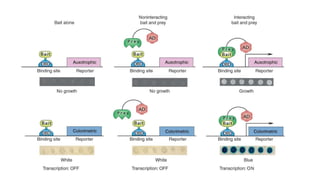
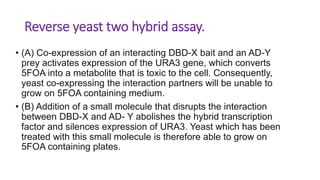
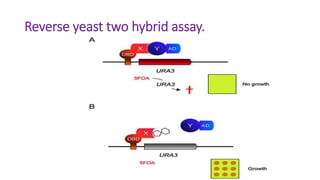
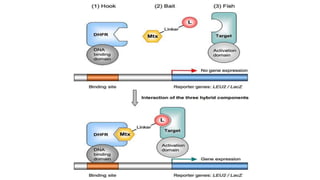
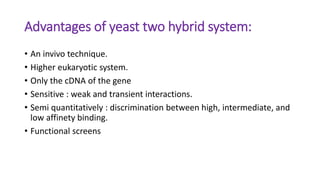
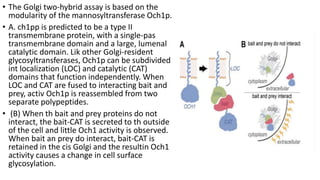
The document discusses the yeast two-hybrid system, which is used to detect protein-protein interactions. It involves fusing a bait protein to a DNA-binding domain and a prey protein to an activation domain. If the bait and prey interact, it brings the domains together and activates transcription of a reporter gene. This allows detection of interactions on selective media or through colorimetric assays. The document covers the basic principles, experimental methods, potential issues like false positives, and modifications like the yeast three-hybrid system.