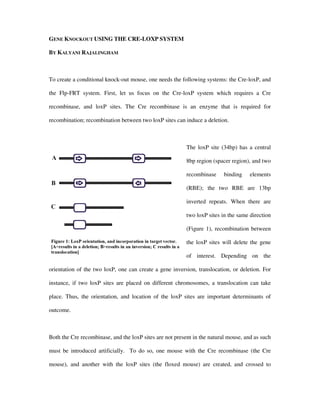
This document describes the Cre-loxP system for creating conditional gene knockouts in mice. The system involves introducing the Cre recombinase enzyme and loxP sites, which are not naturally present in mice. LoxP sites are inserted on either side of the gene to be knocked out in one "floxed" mouse line. A separate "Cre mouse" line is created that expresses Cre recombinase in a tissue-specific manner. Crossing the two mouse lines results in Cre recombinase recombining the loxP sites and deleting the gene of interest, but only in the tissues where Cre is expressed, allowing conditional rather than complete knockout of the gene.