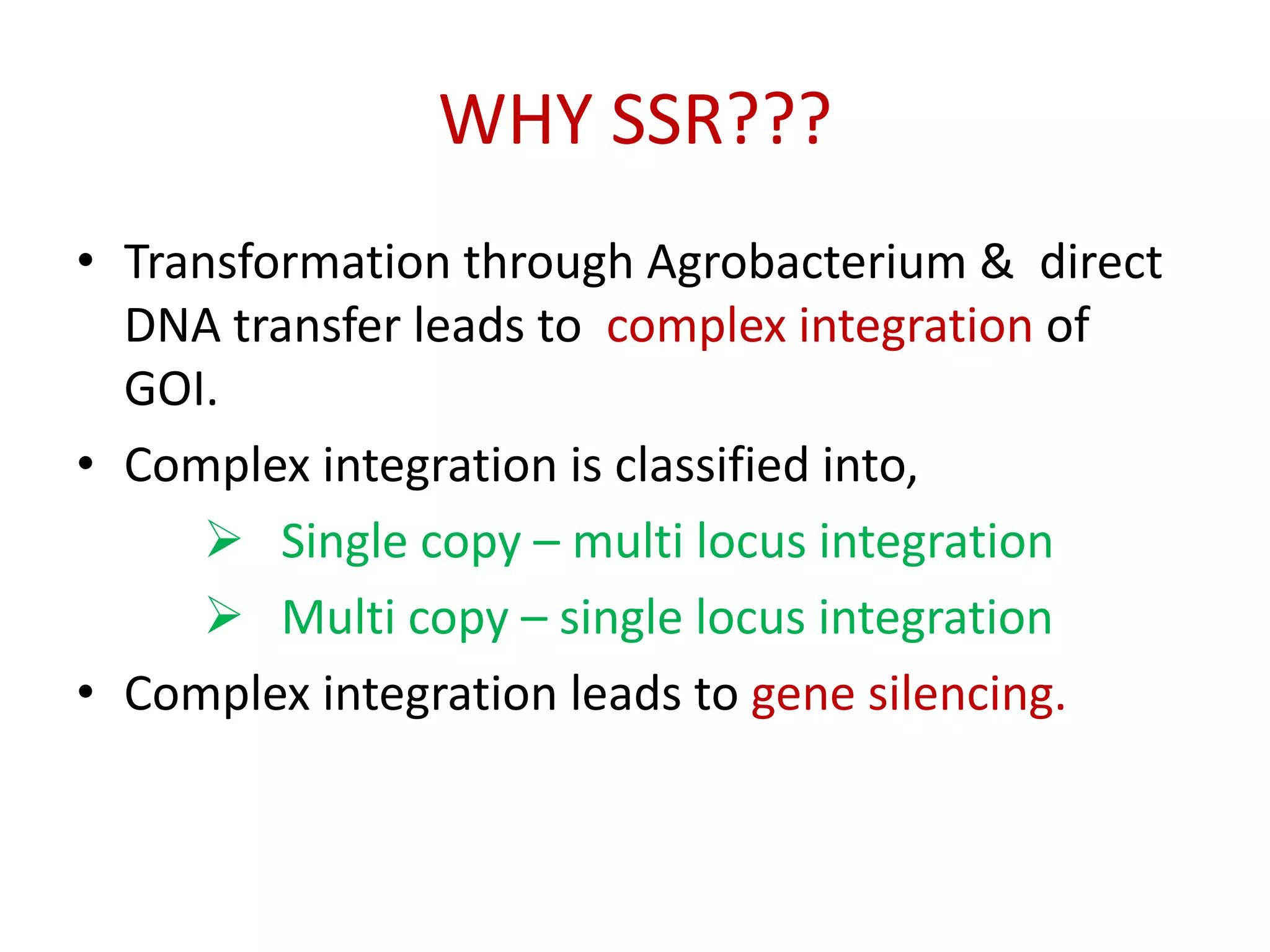
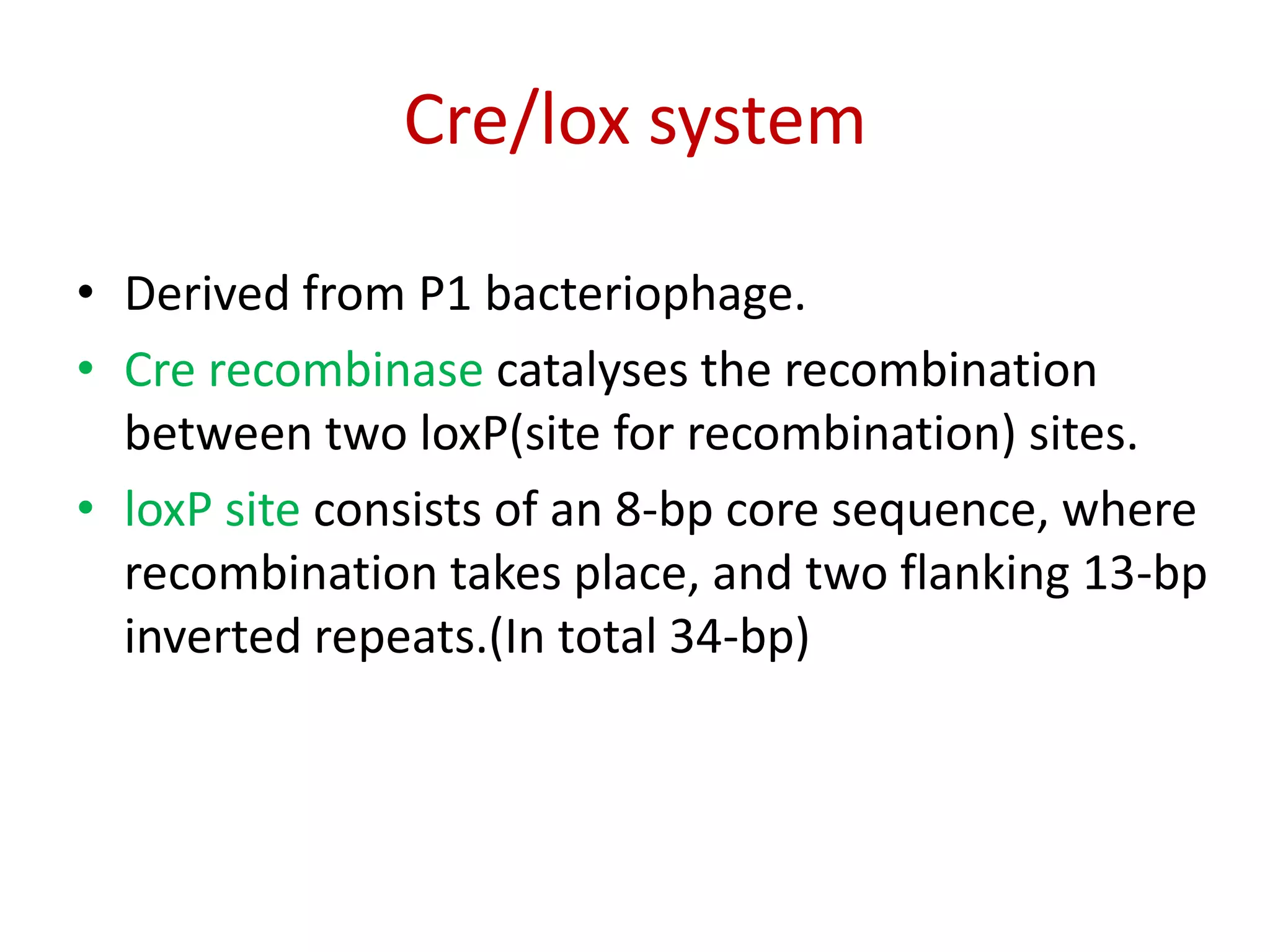
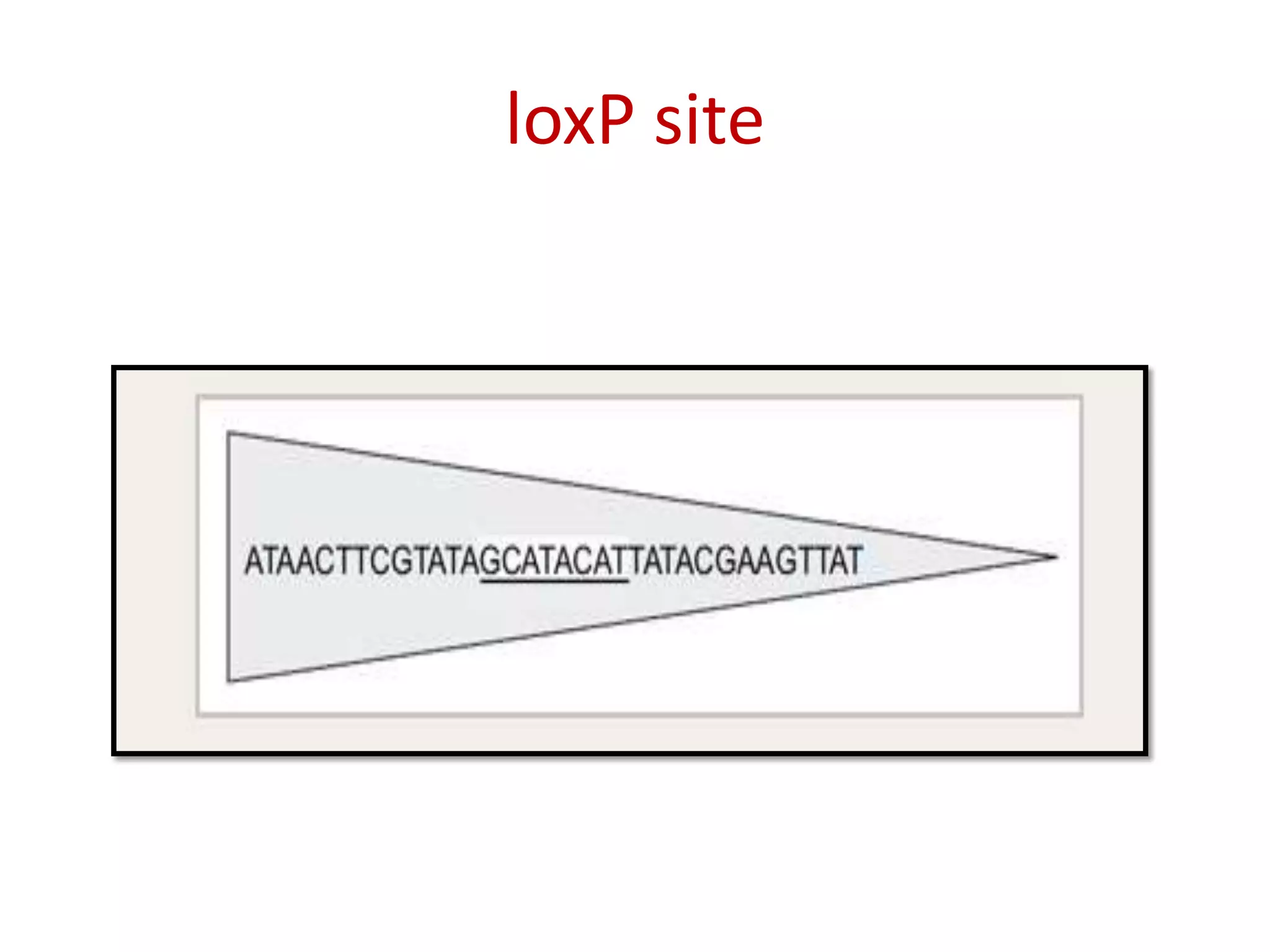
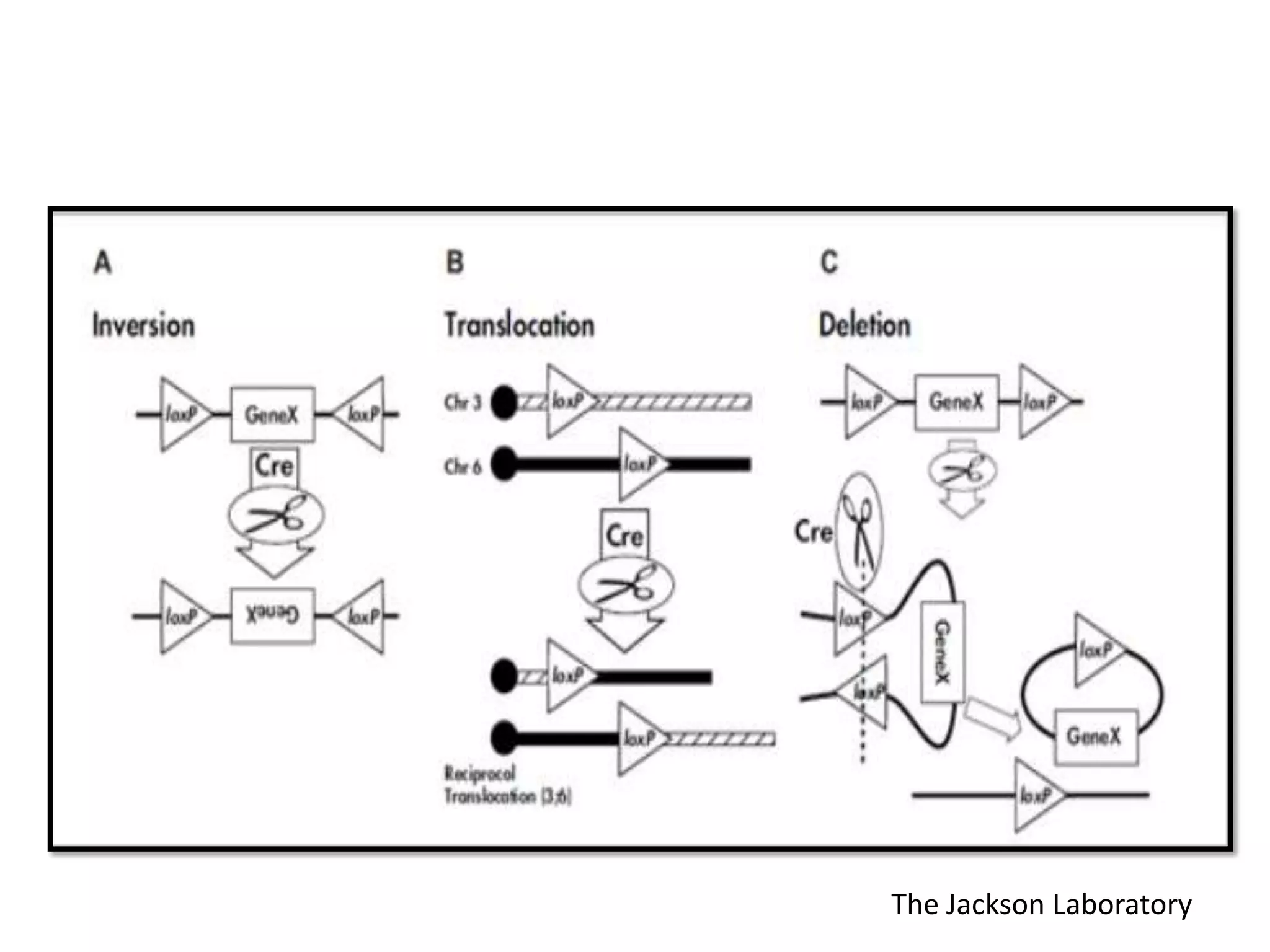
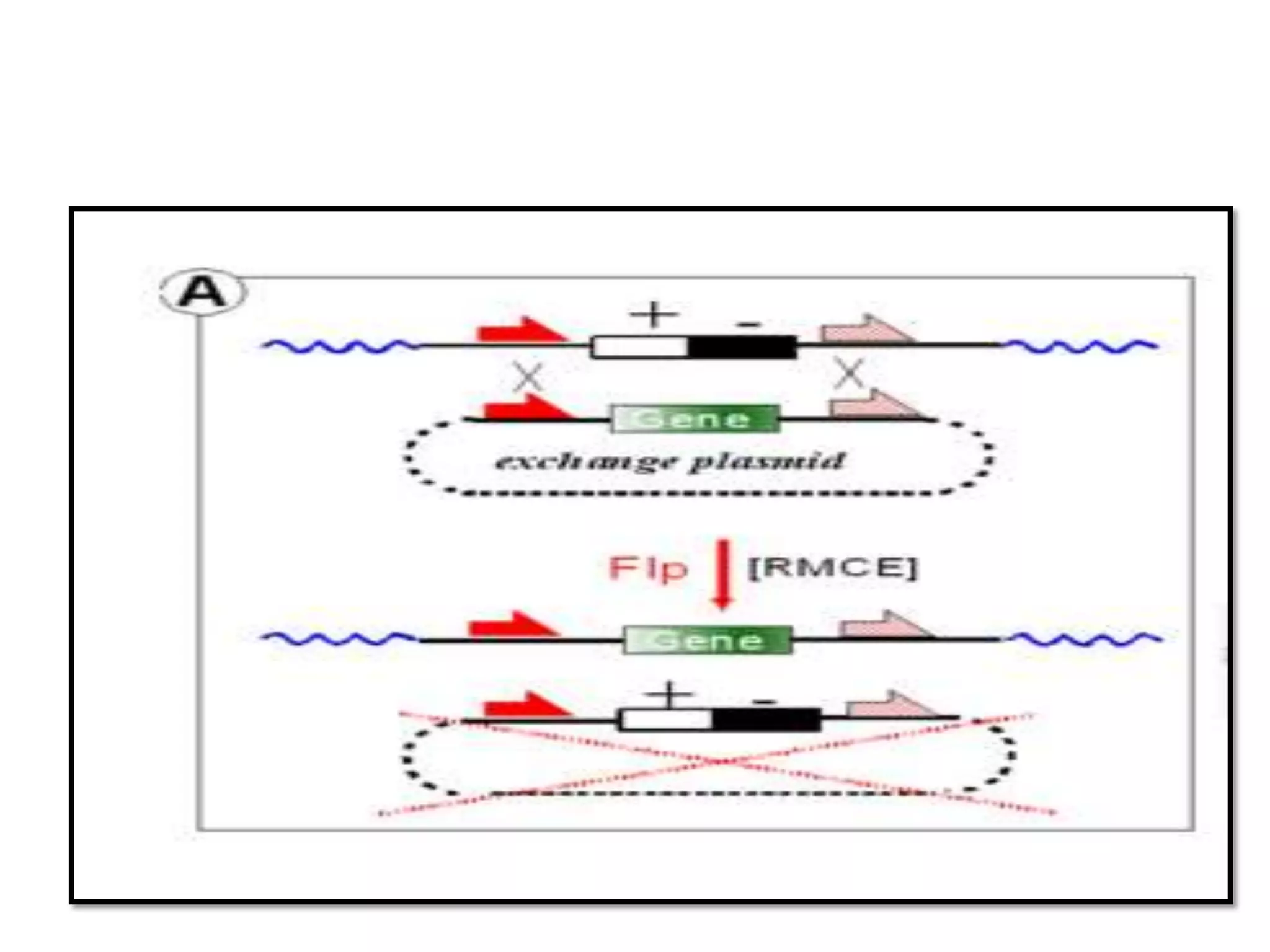
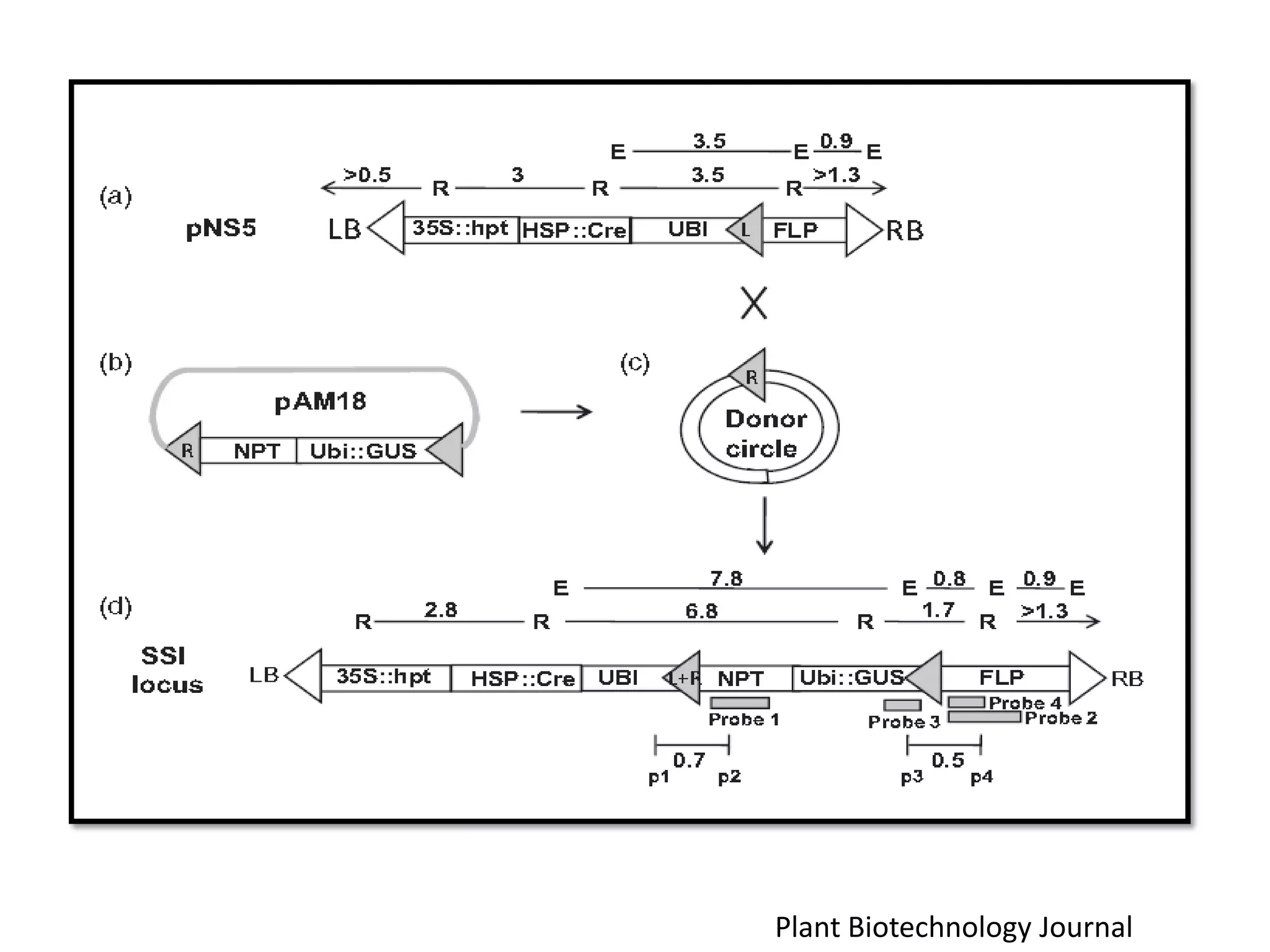
Site-specific recombination (SSR) systems like Cre/lox and FLP/FRT allow for precise integration of genes of interest (GOI) into plant genomes compared to traditional transformation methods. The Cre/lox system uses the Cre recombinase to catalyze recombination between two loxP sites, while the FLP/FRT system uses FLP recombinase to catalyze recombination between two FRT sites. These systems allow for single-copy, single-locus integration of GOIs without disrupting functional genes. However, constitutive expression of Cre and FLP recombinases can cause unwanted genomic changes. Current research focuses on engineering these recombinases for improved efficiency and versatility of SSR for transgenic plant