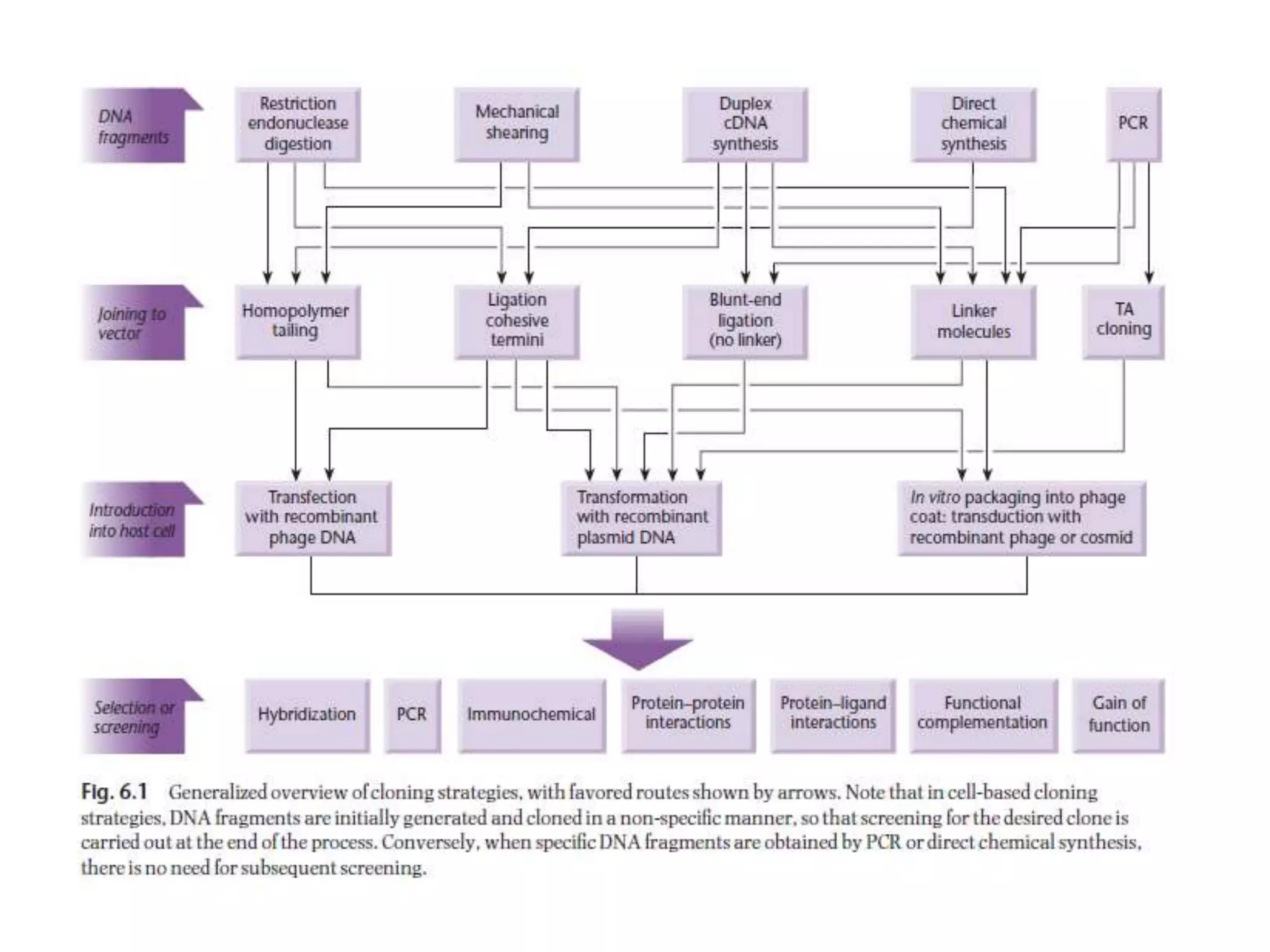
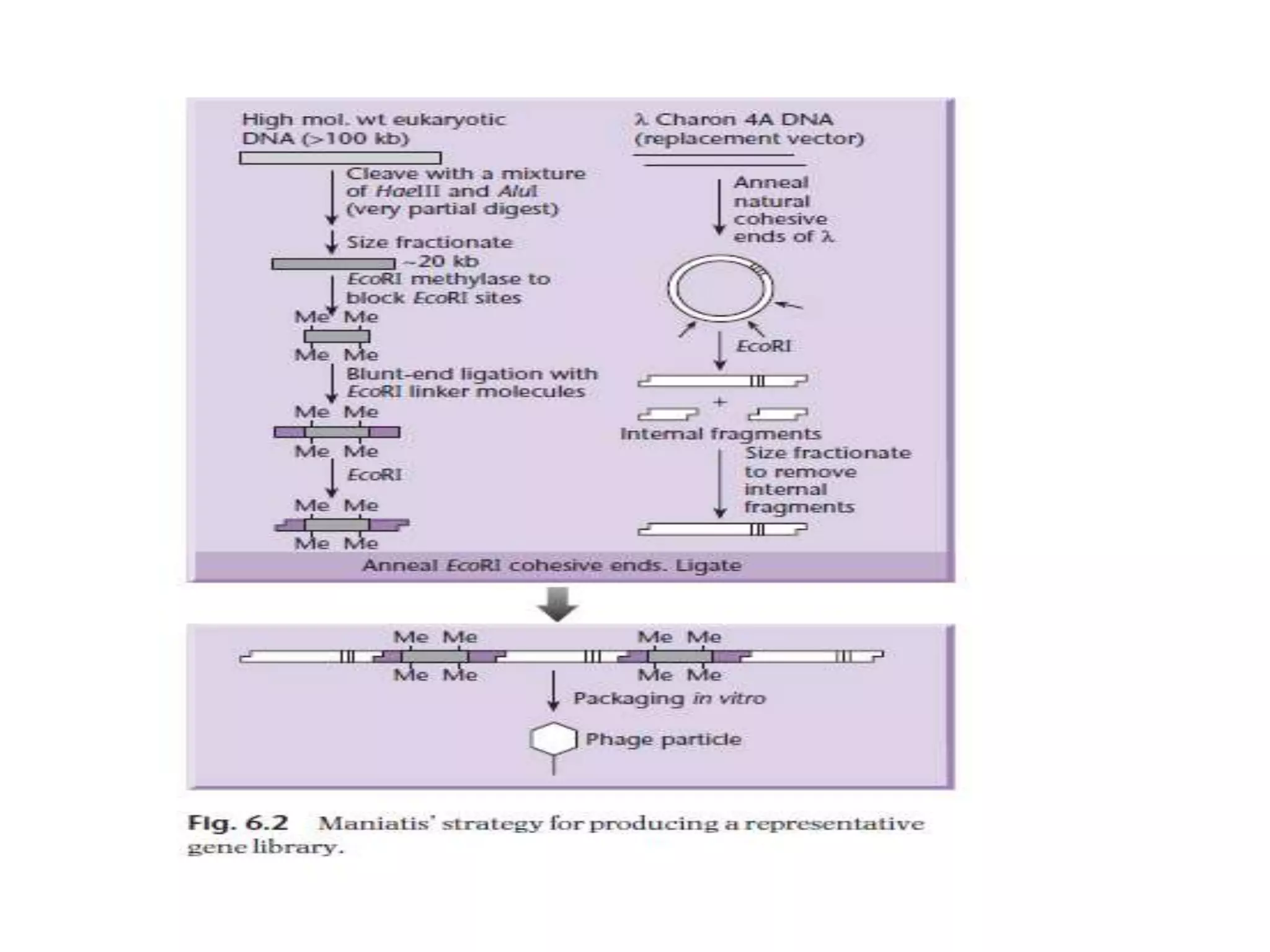
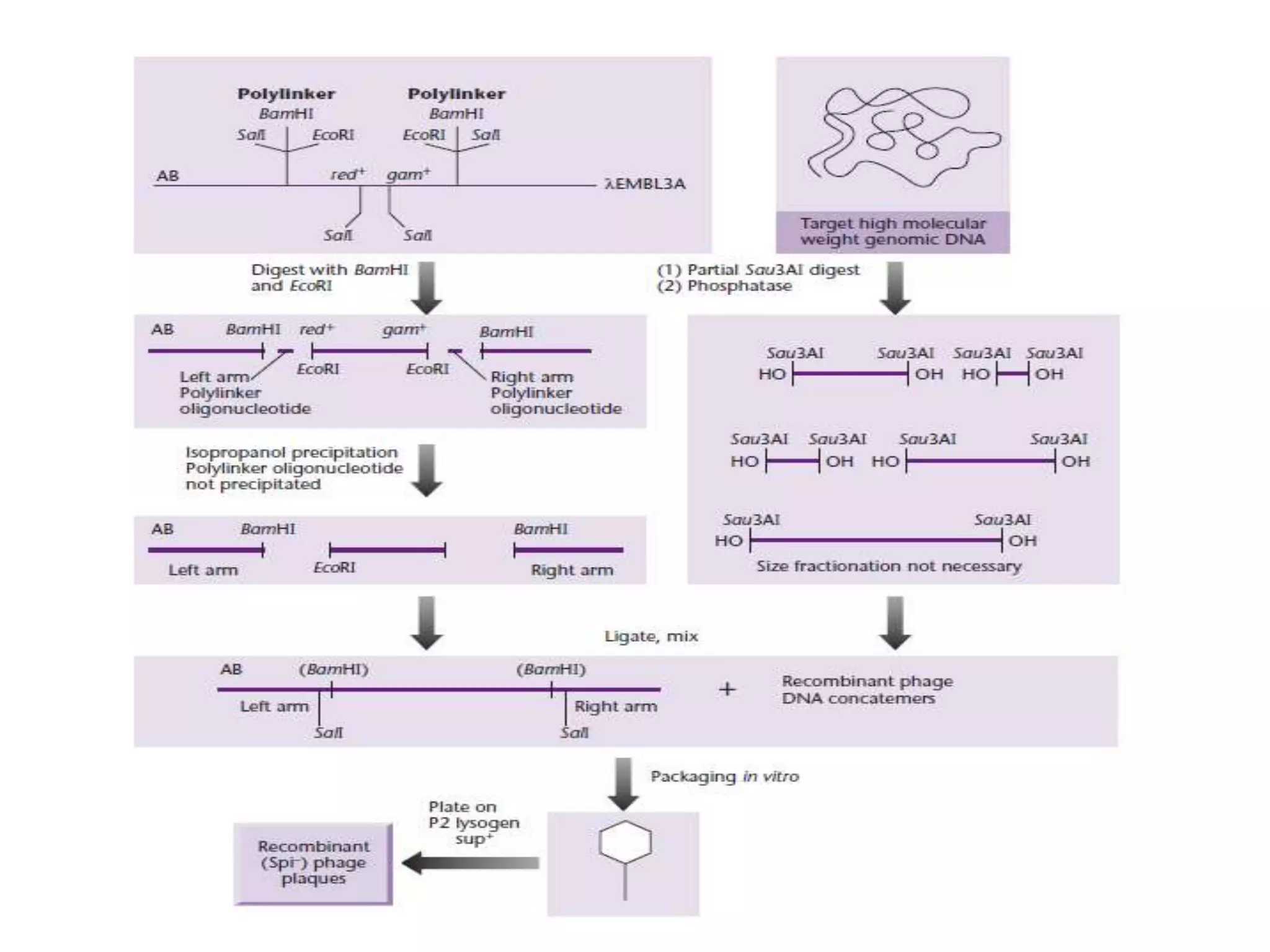
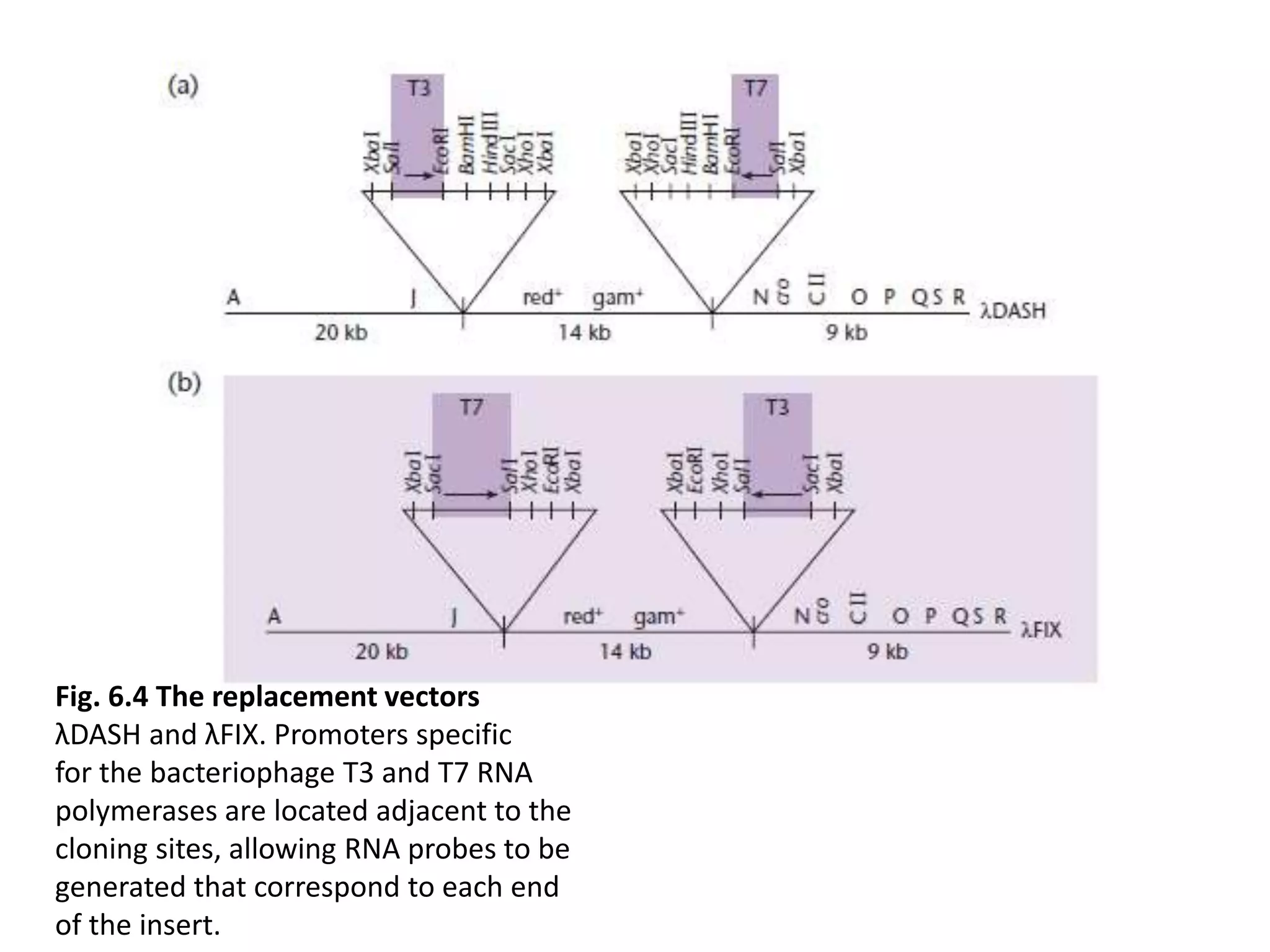
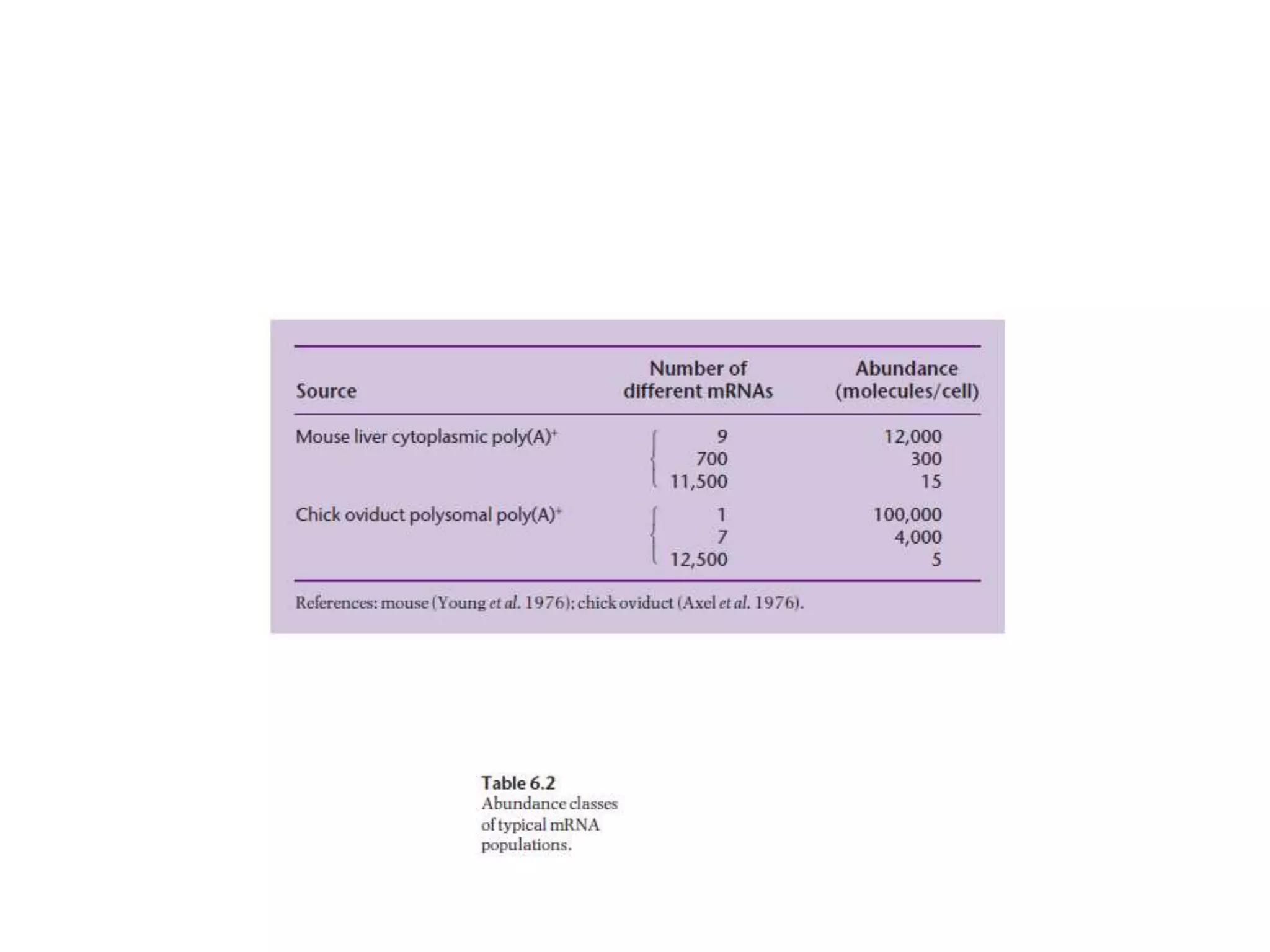
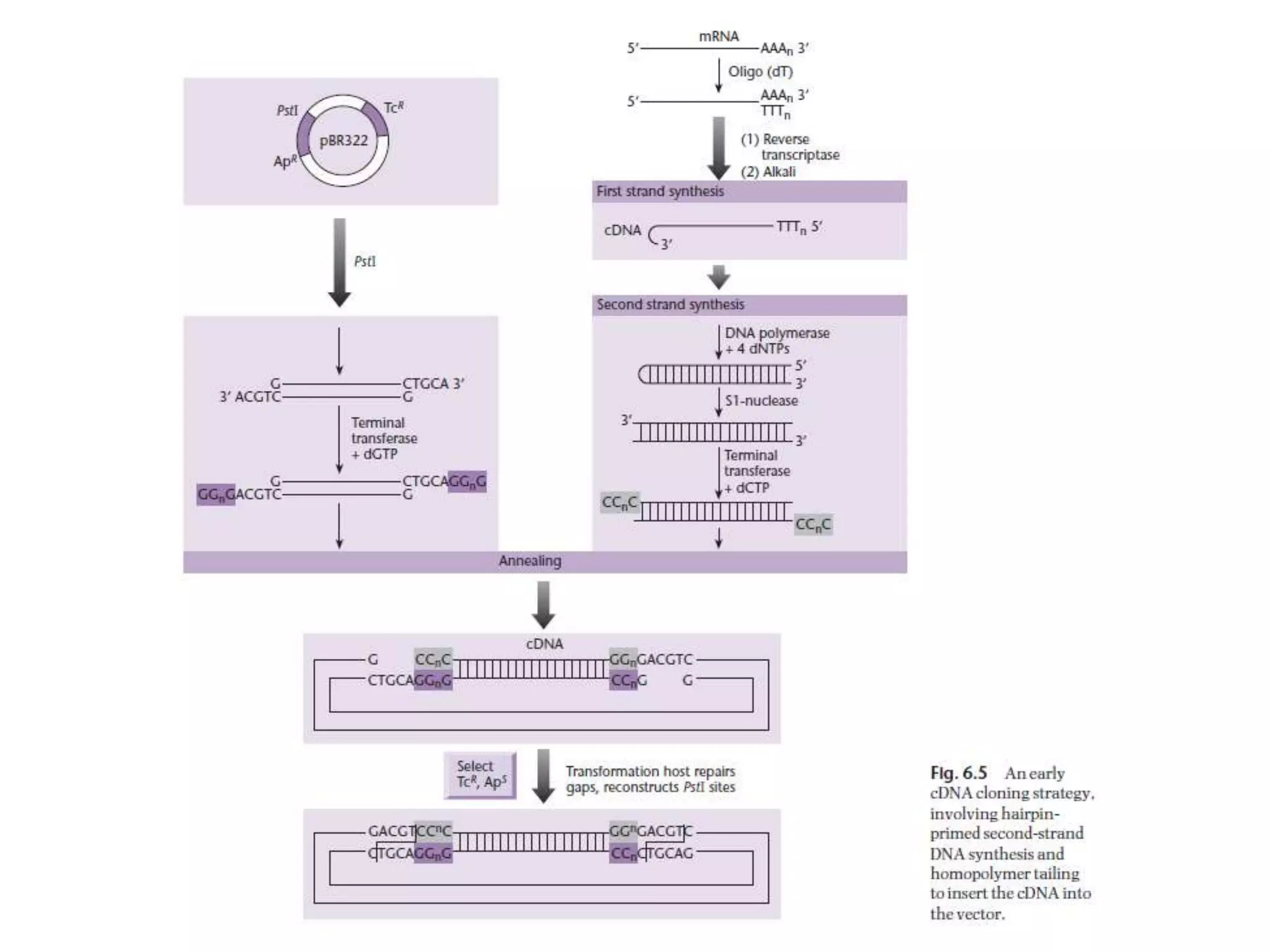
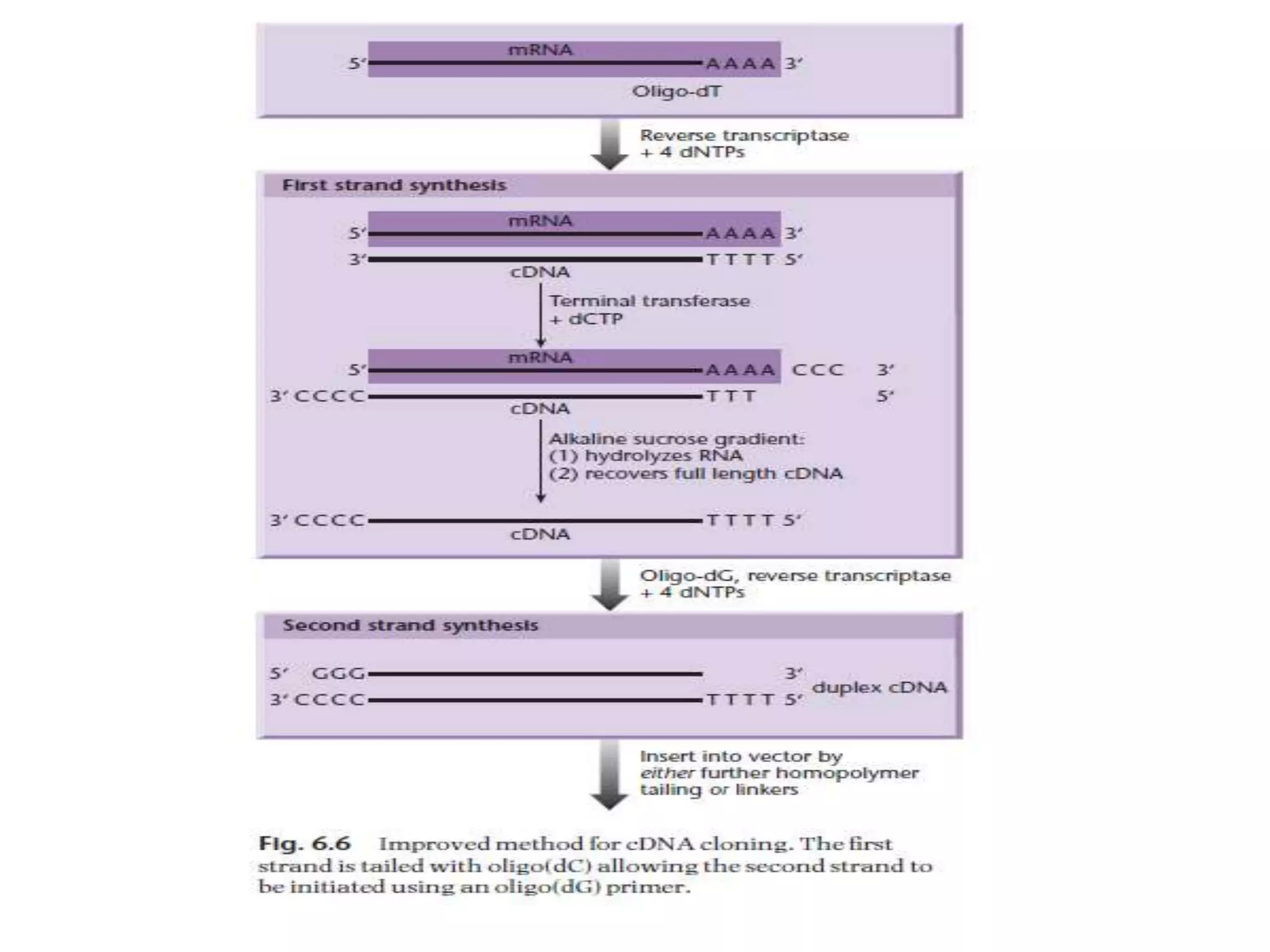
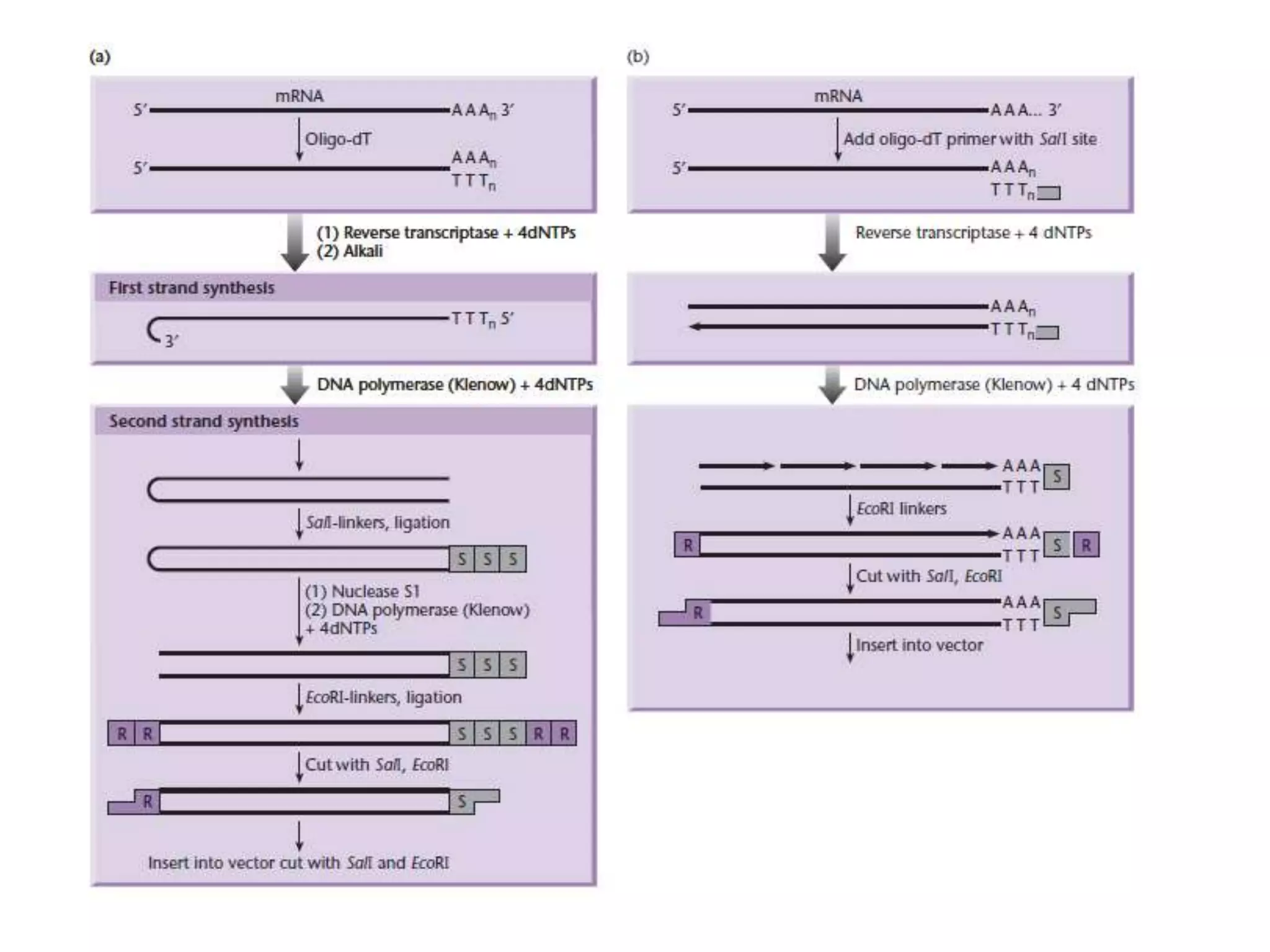
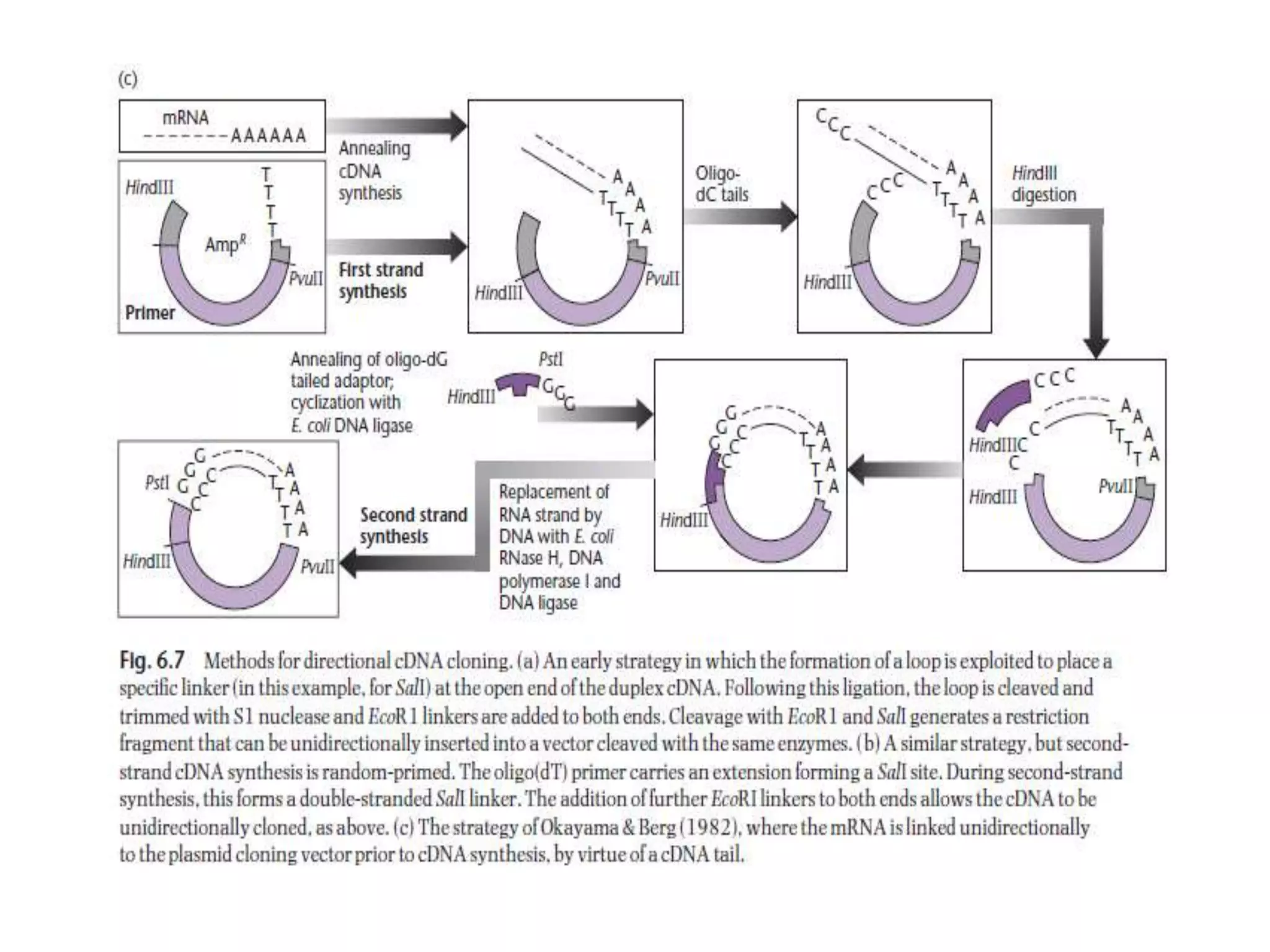
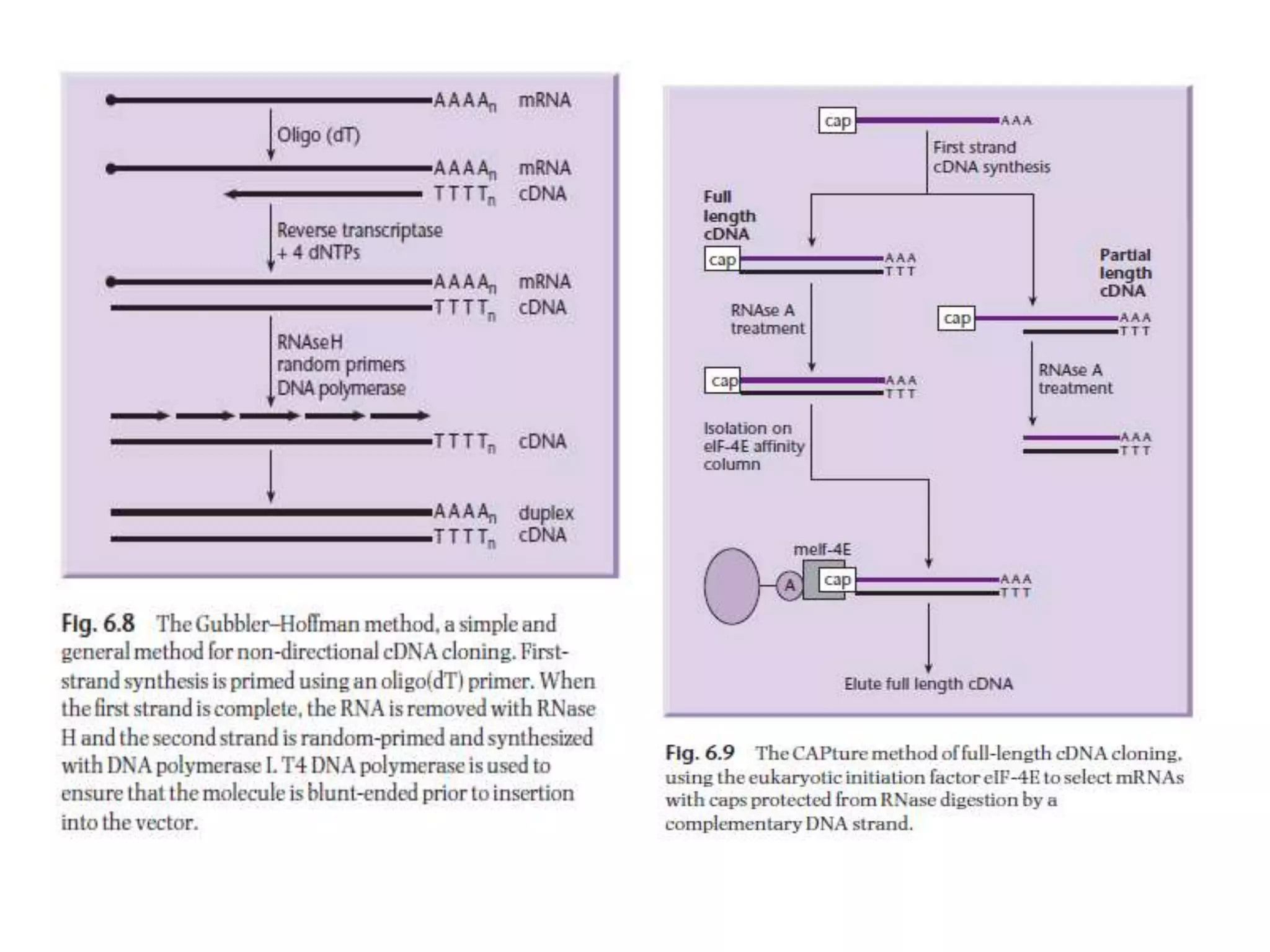
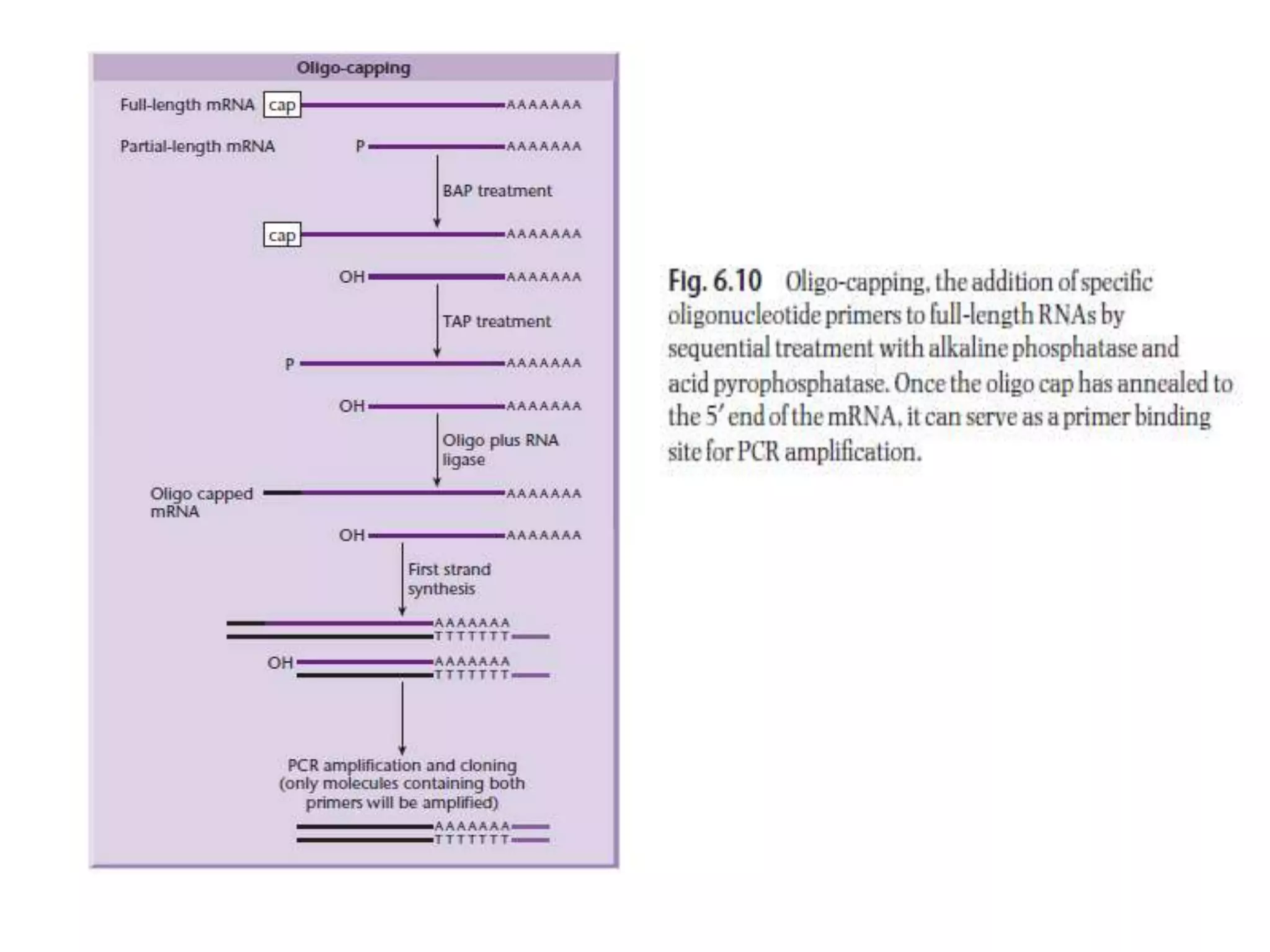
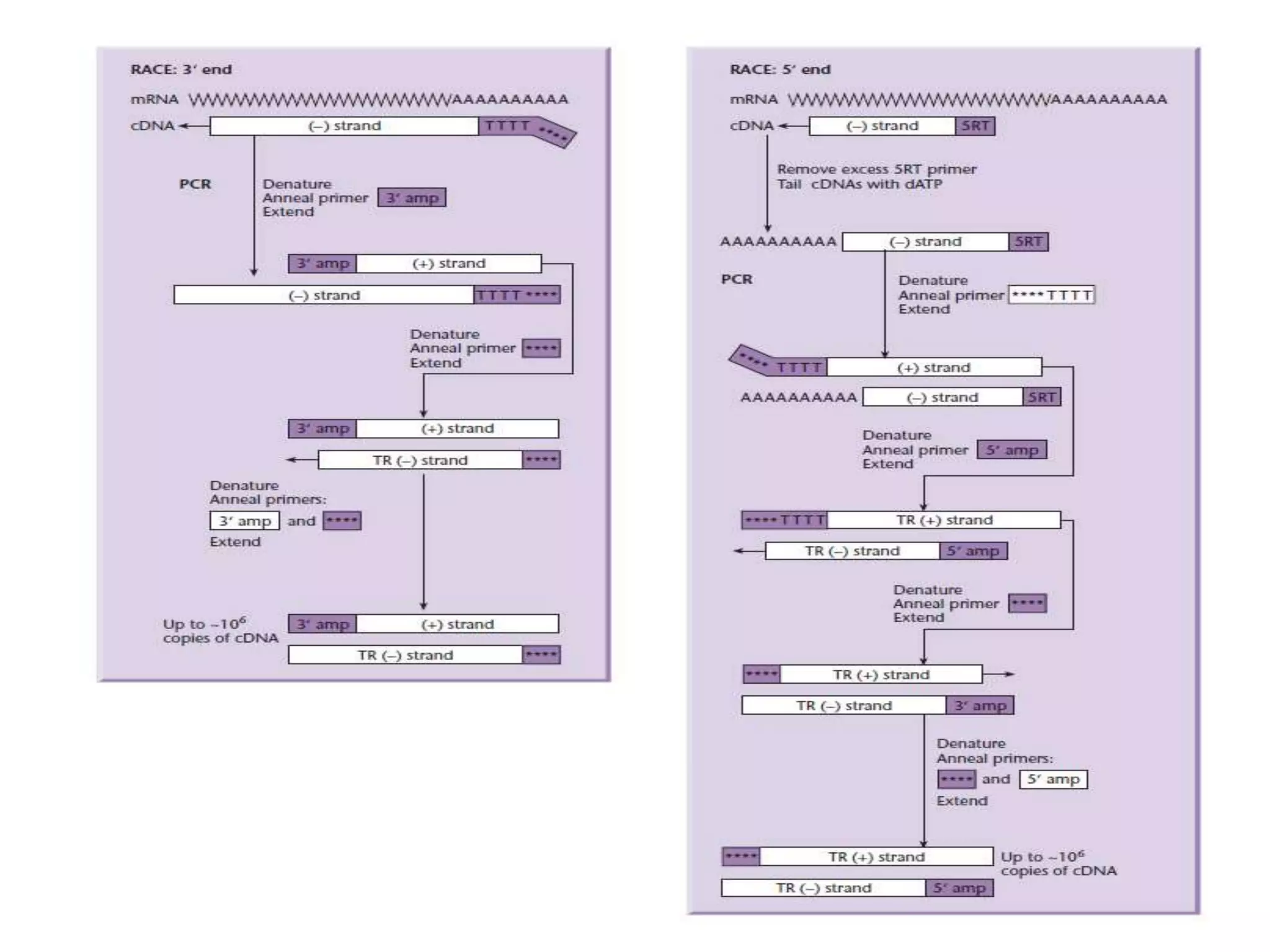
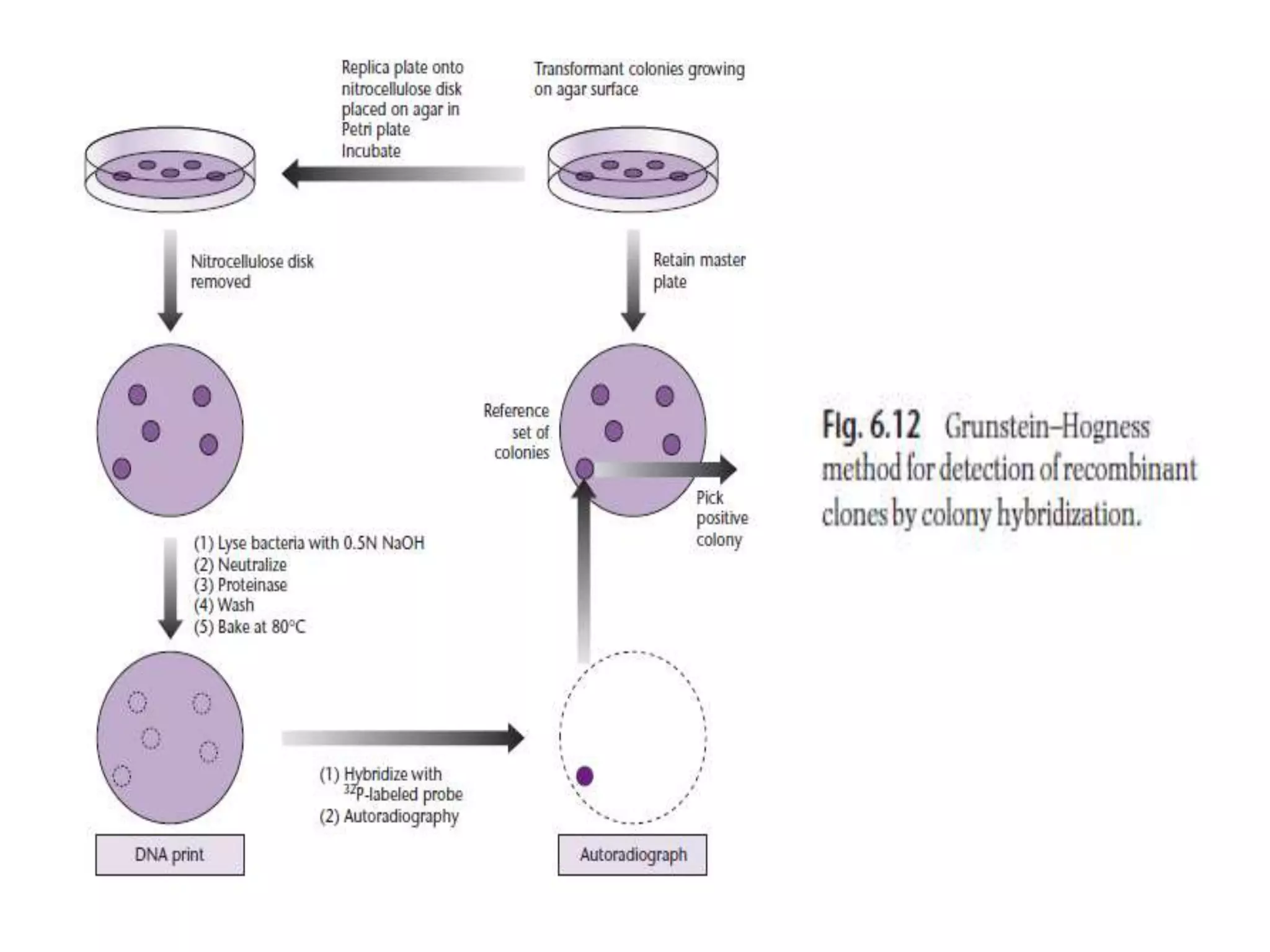
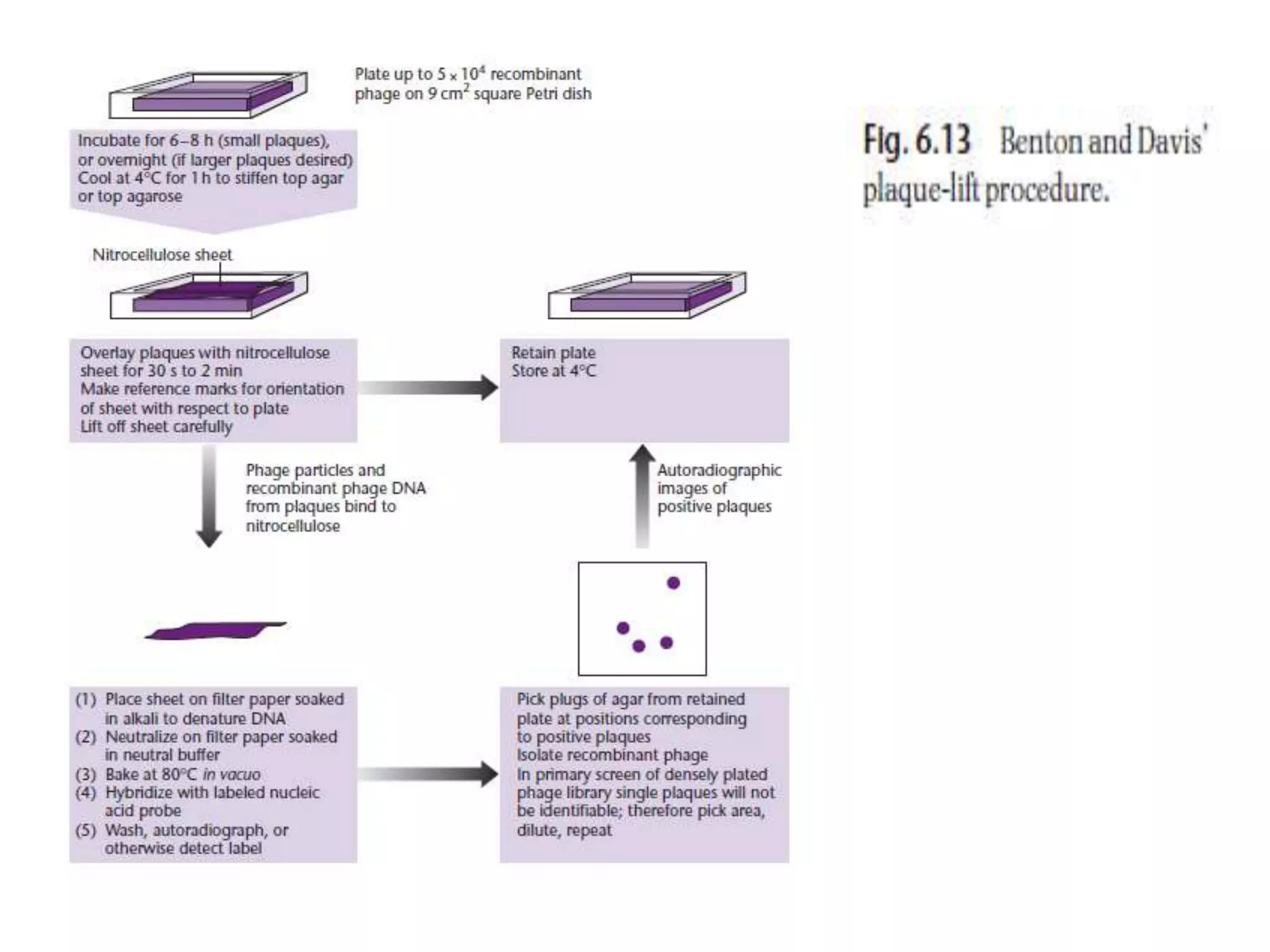
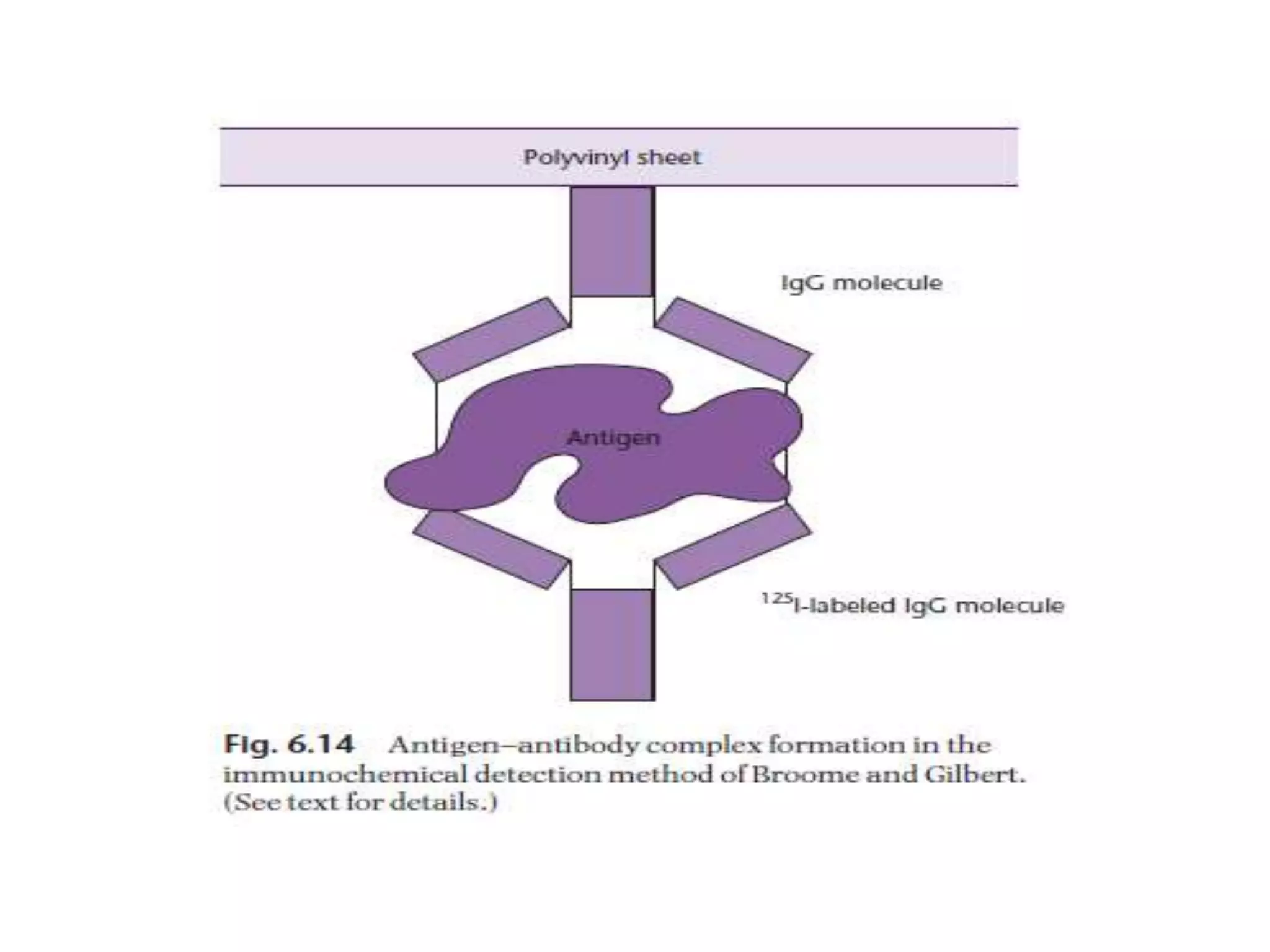
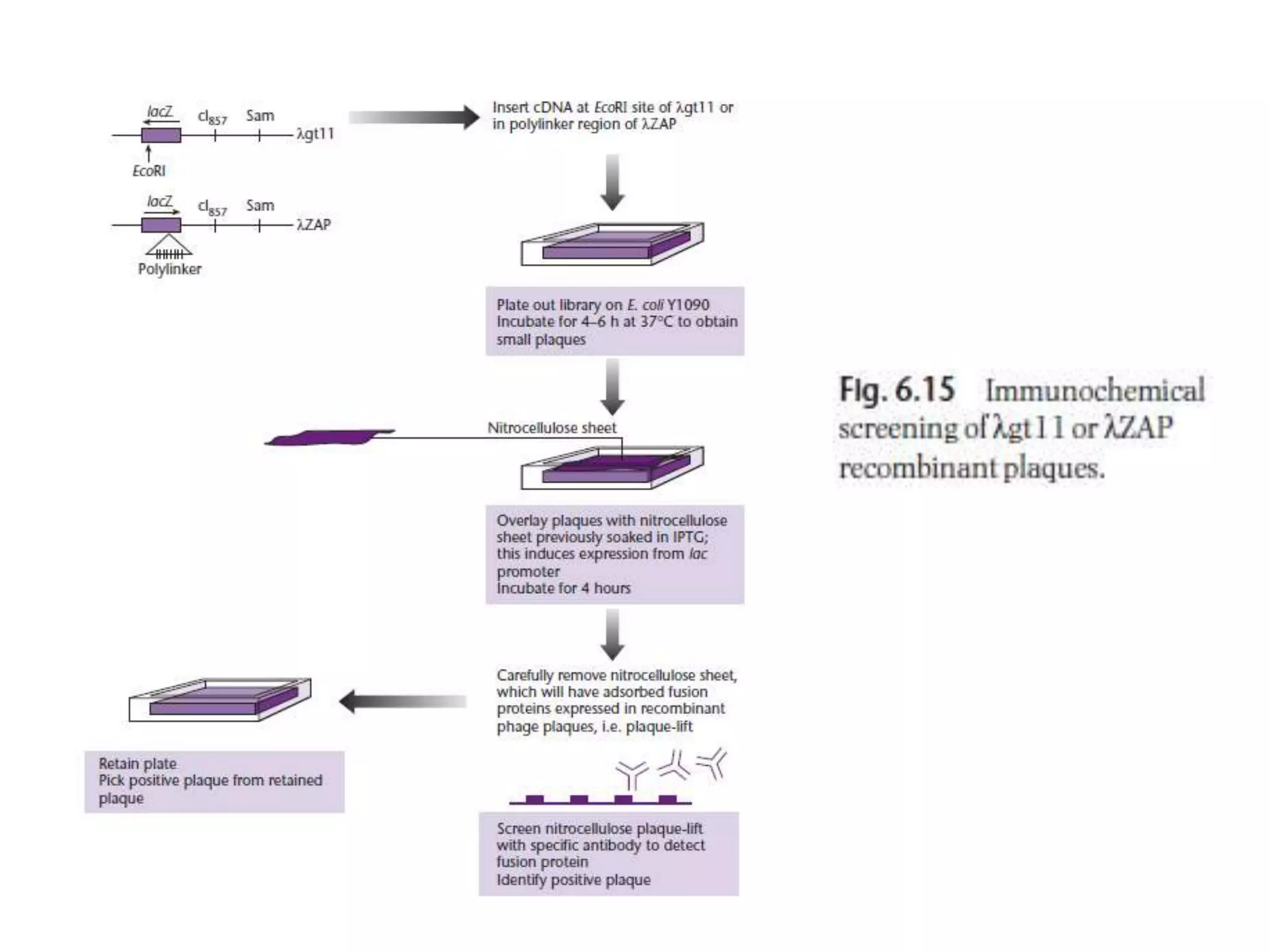
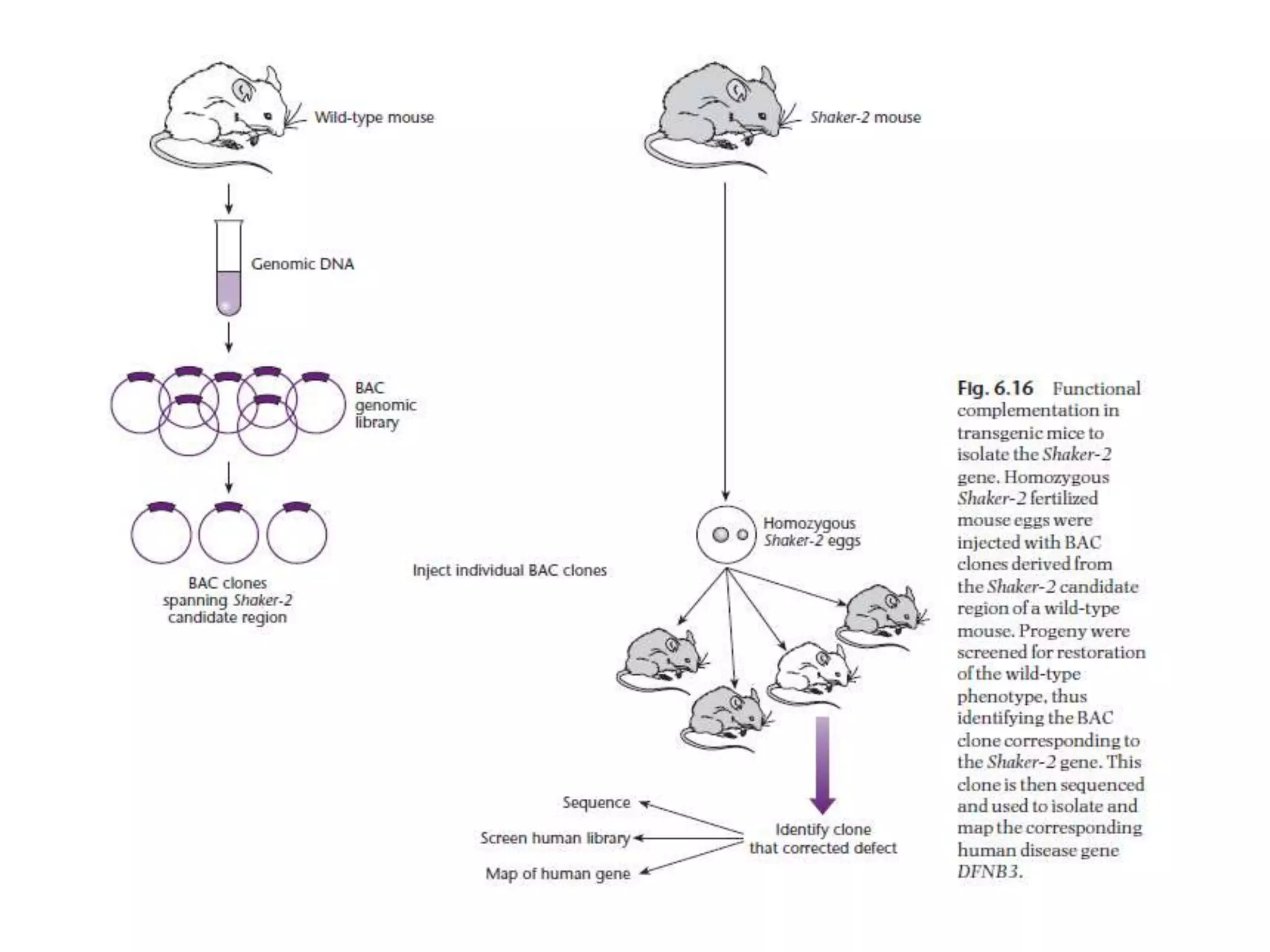
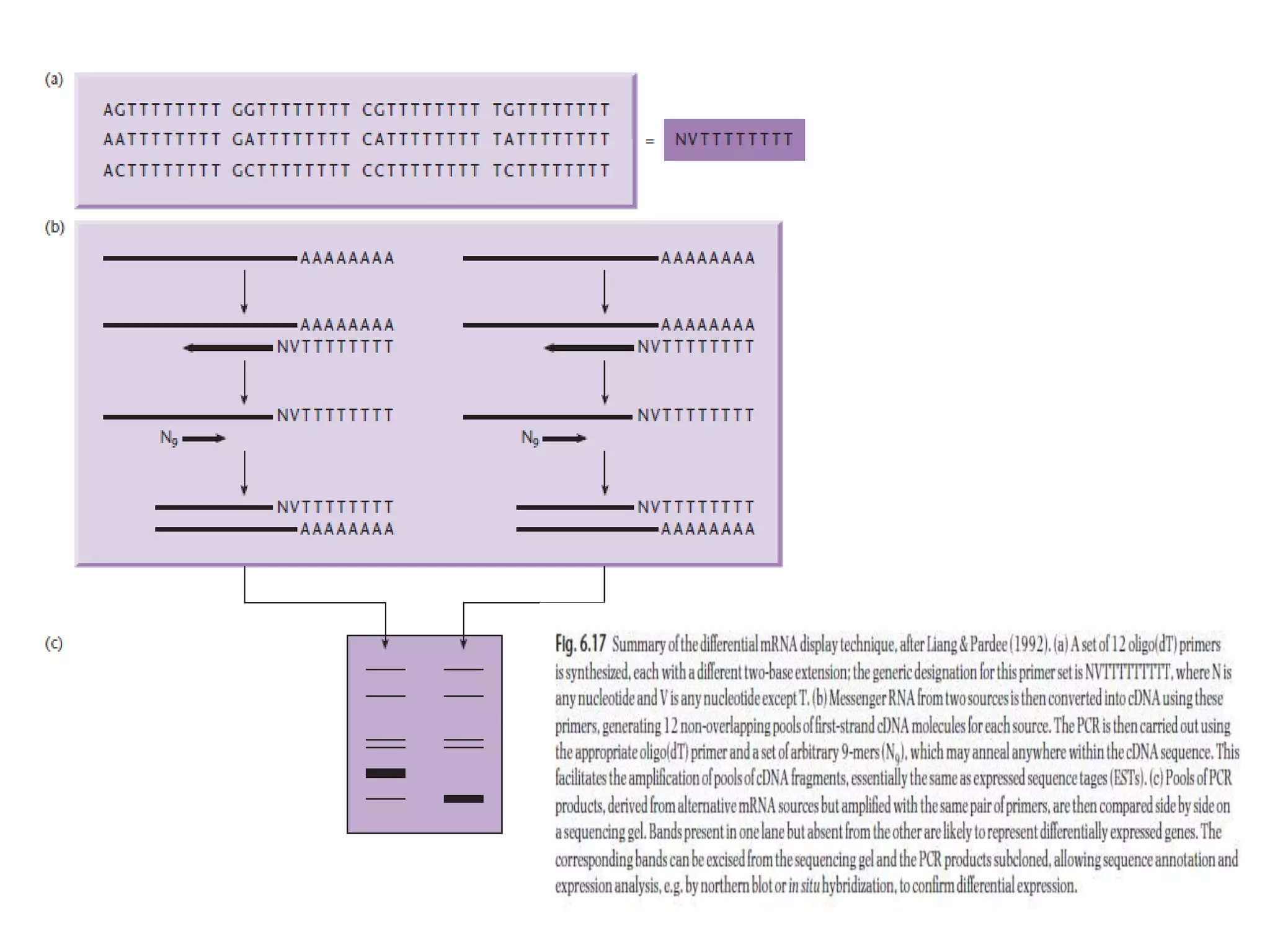
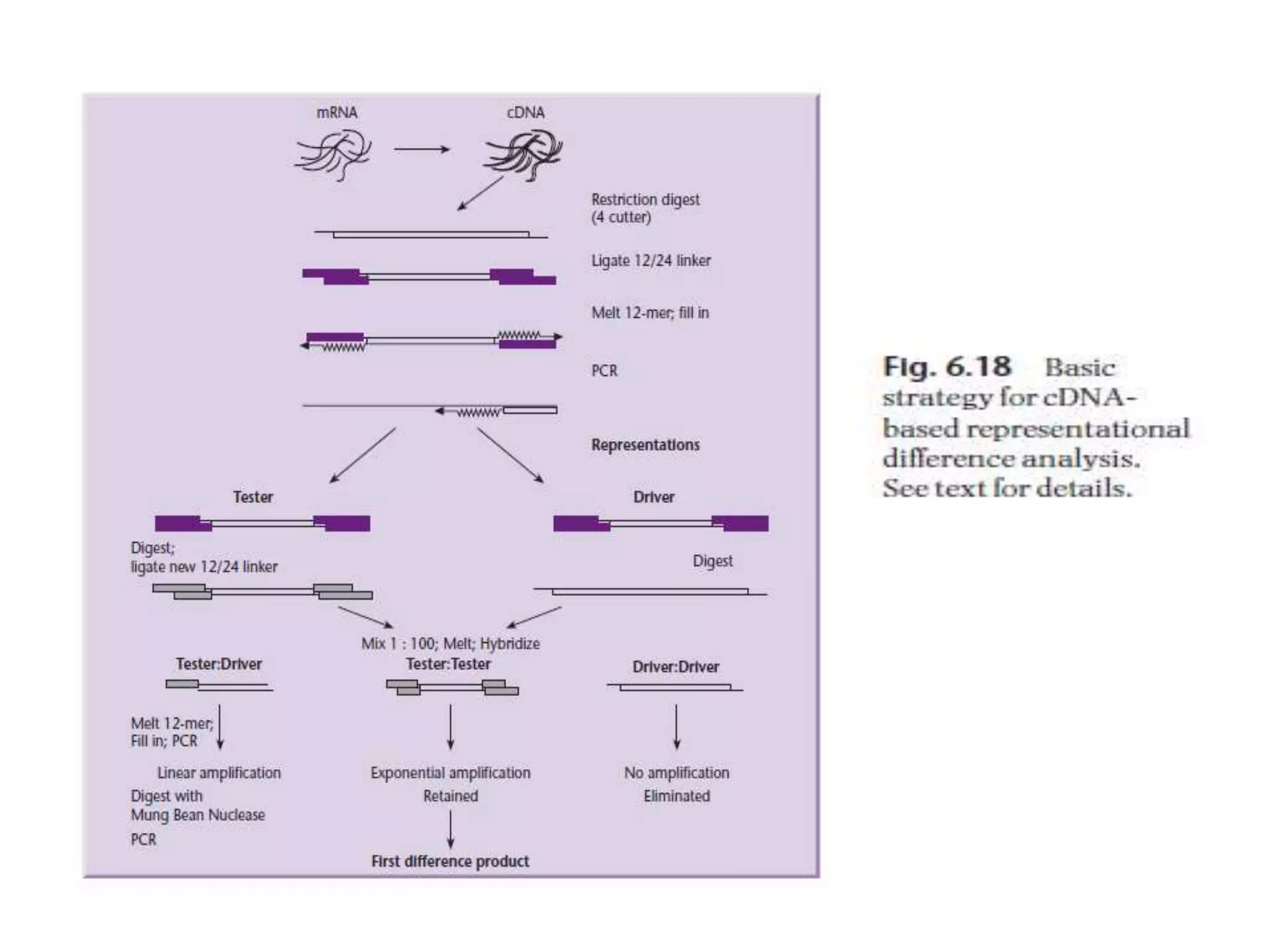
The document describes a strategy for creating a genomic library using restriction enzymes. A partial digest is performed using two enzymes that cut frequently, producing random overlapping fragments around 20kb in size. These fragments are inserted into replacement vectors and packaged in vitro to create an almost completely representative library. Alternatively, a single frequent cutting enzyme like Sau3AI can be used, which creates fragments that can be readily inserted into lambda replacement vectors.