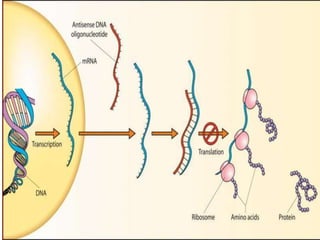
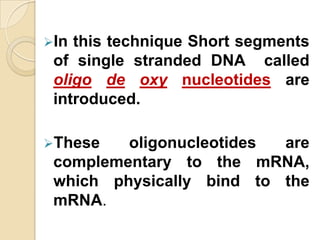
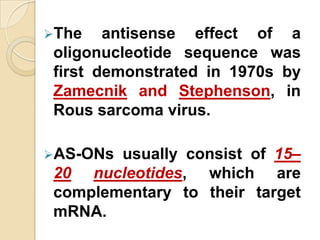
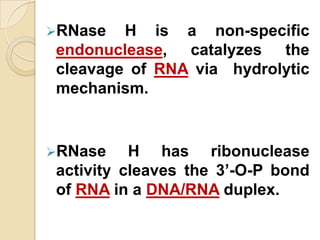
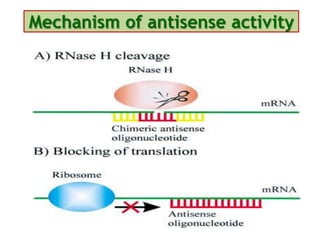
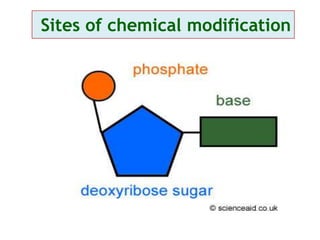
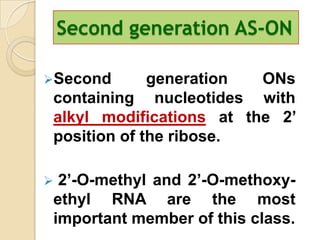
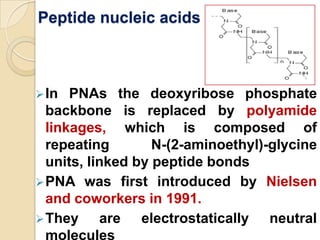
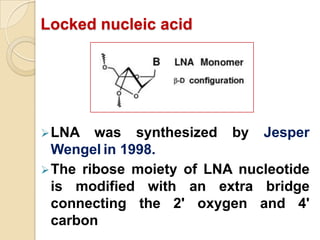
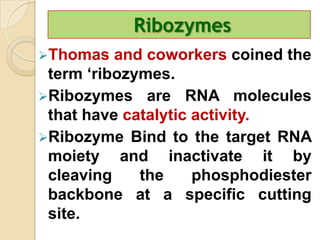
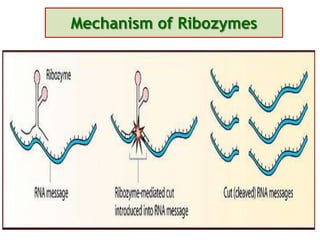
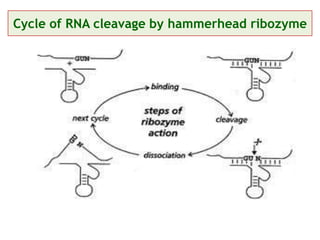
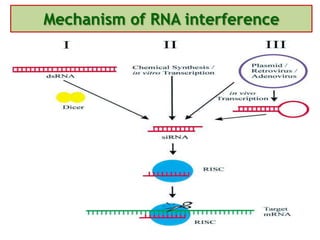
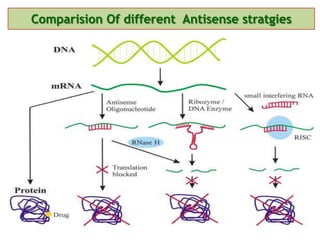
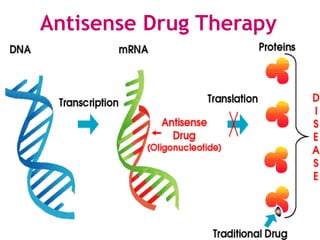
Antisense technology uses short DNA sequences called oligonucleotides that are complementary to messenger RNA (mRNA) to prevent specific proteins from being synthesized. When introduced into cells, these antisense oligonucleotides bind to their target mRNA through Watson-Crick base pairing, forming RNA-DNA hybrids that are degraded by RNase H enzyme. This prevents translation and expression of the target protein. There are three generations of antisense oligonucleotides that have been developed with improved stability and targeting capabilities, including phosphorothioate, 2'-O-methyl RNA, and locked nucleic acid chemistries. Antisense technology has potential applications in treating diseases like cancer, viral infections, and genetic disorders.