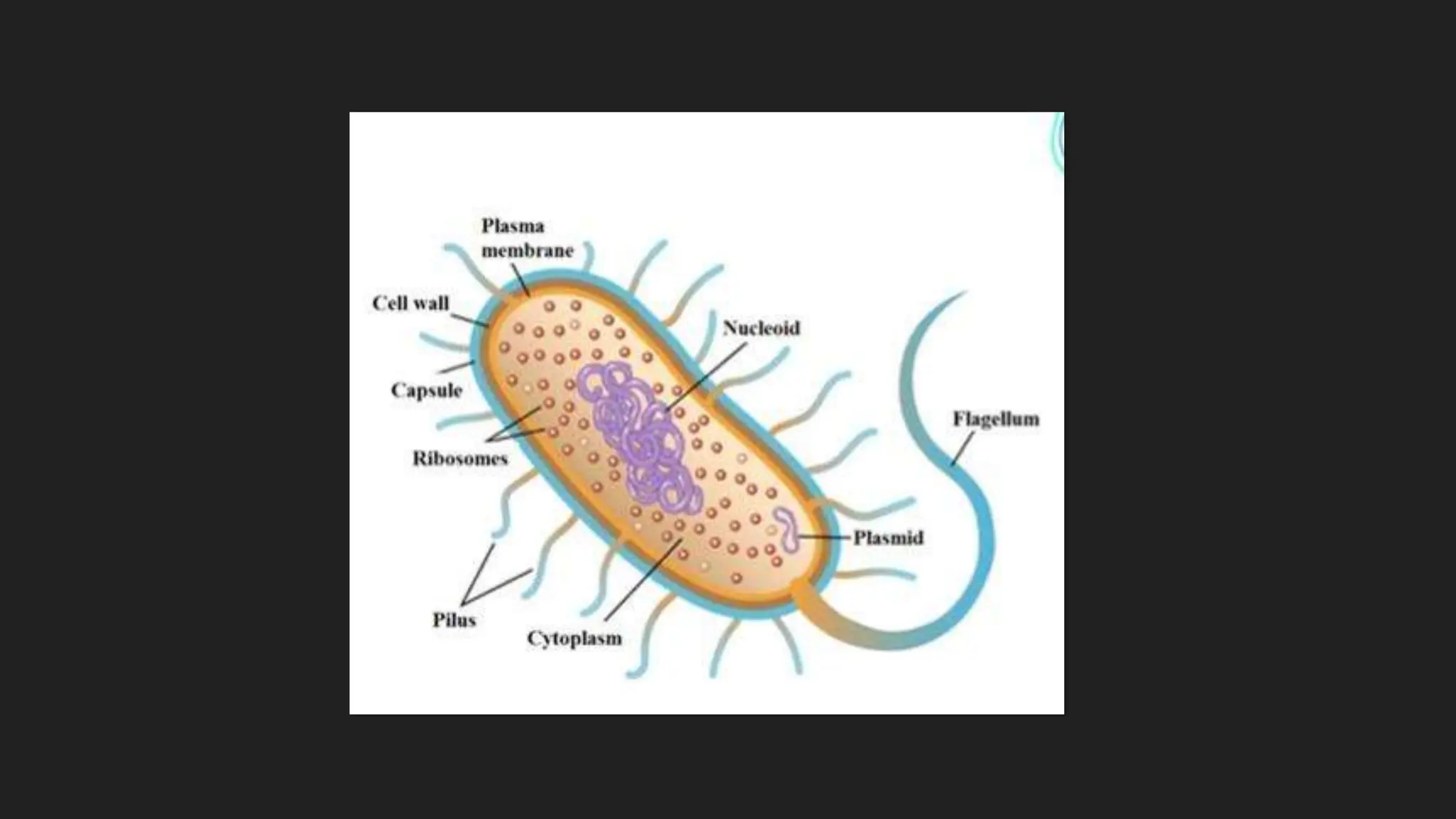
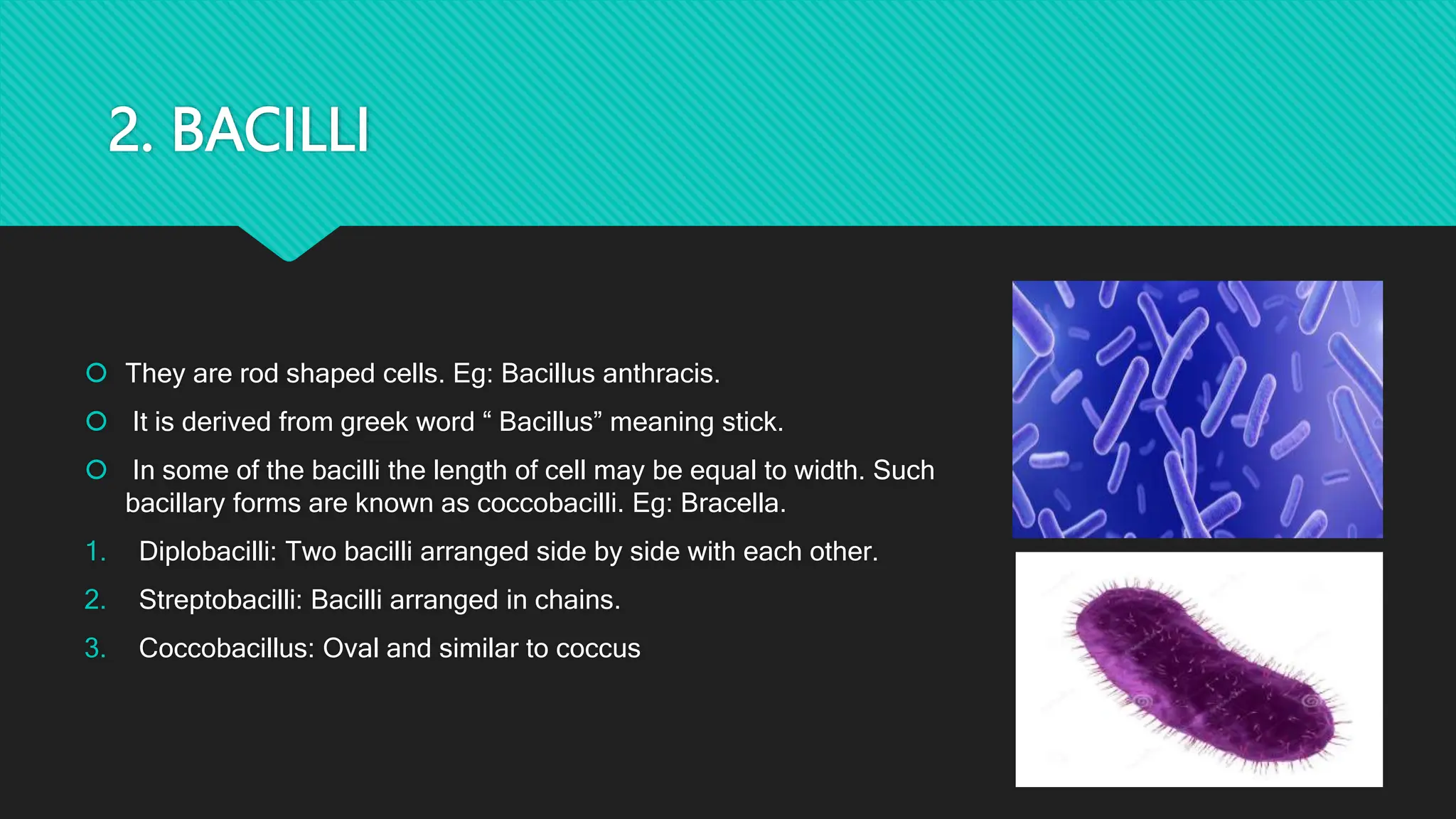
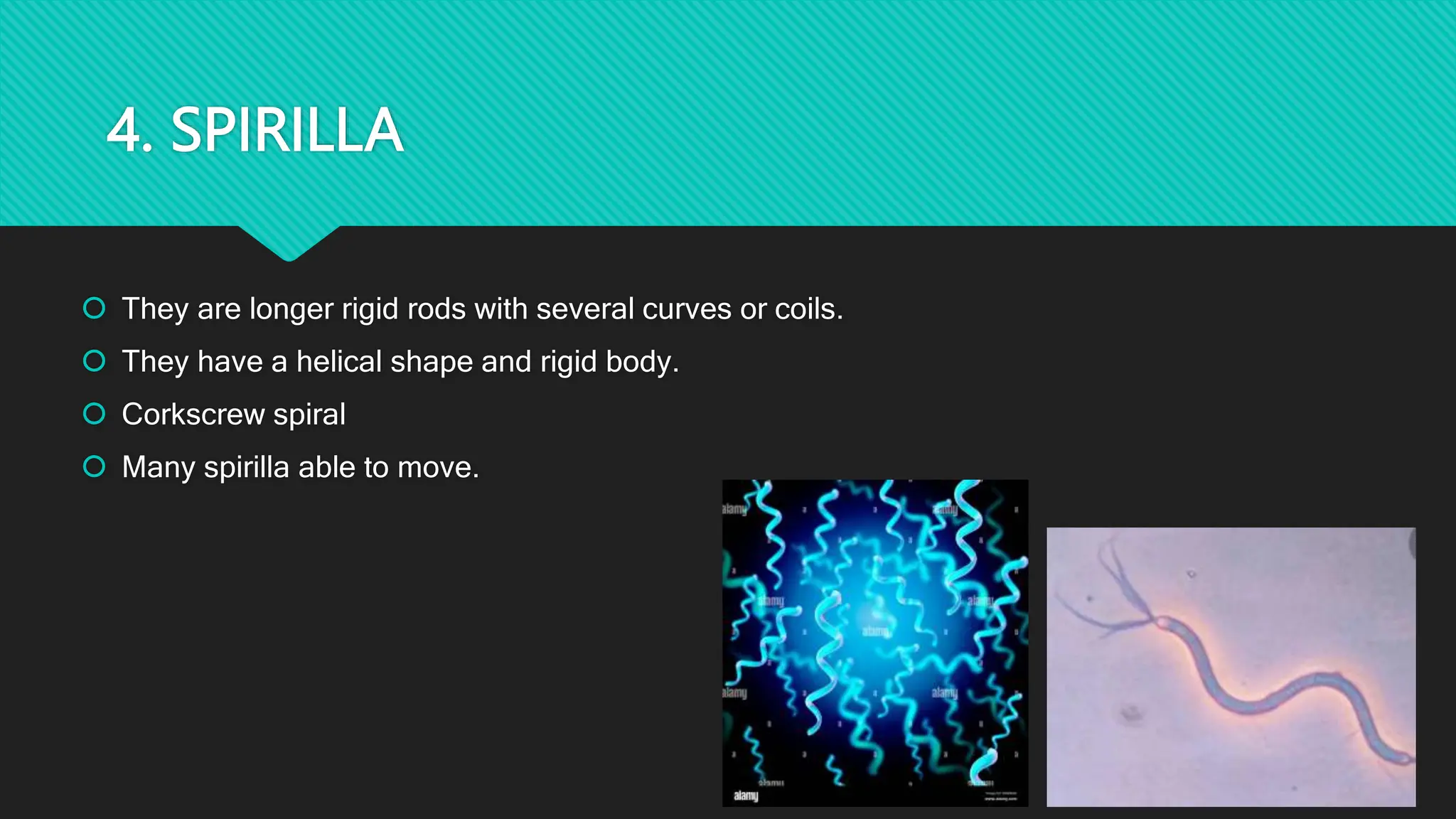
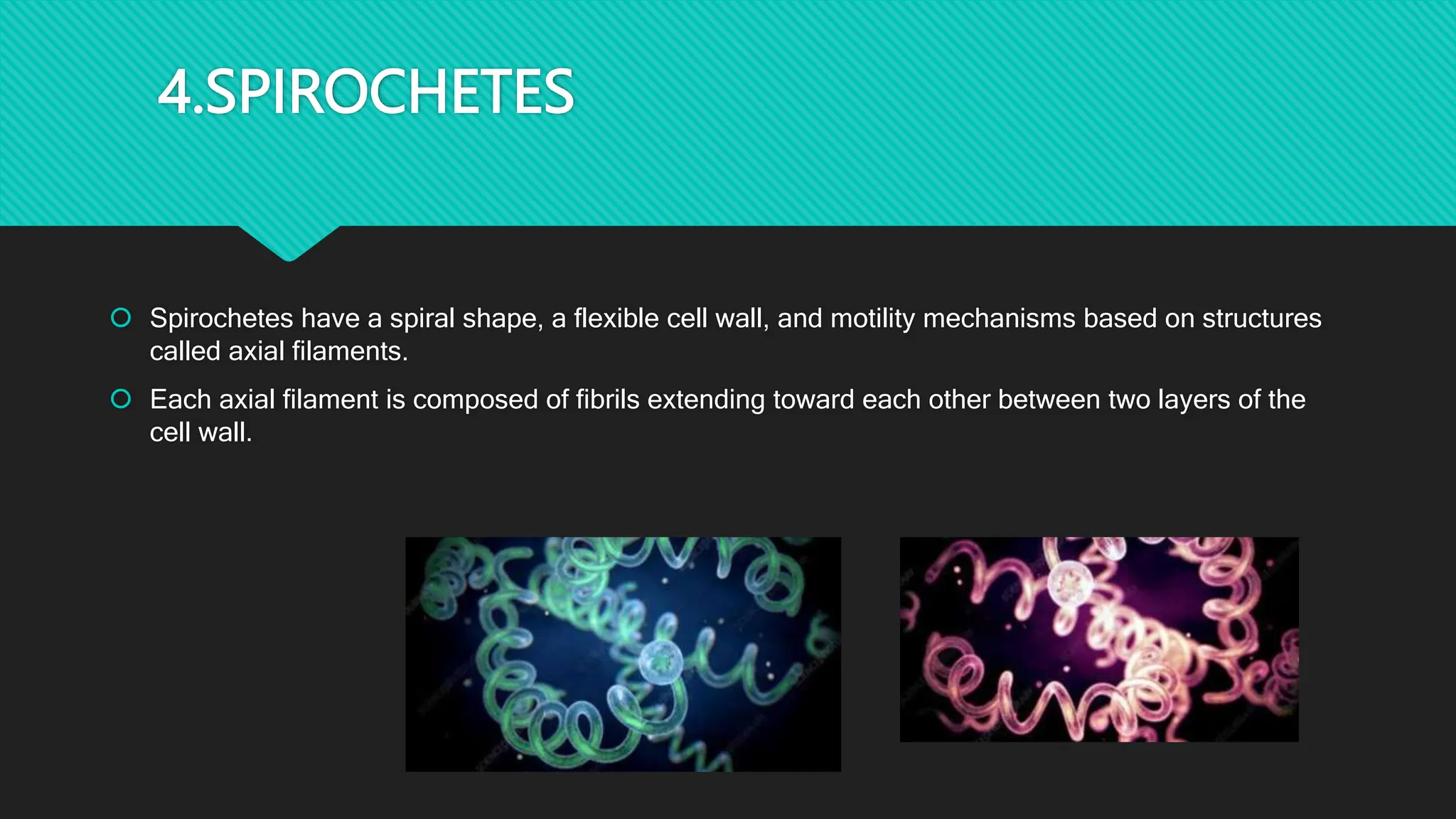
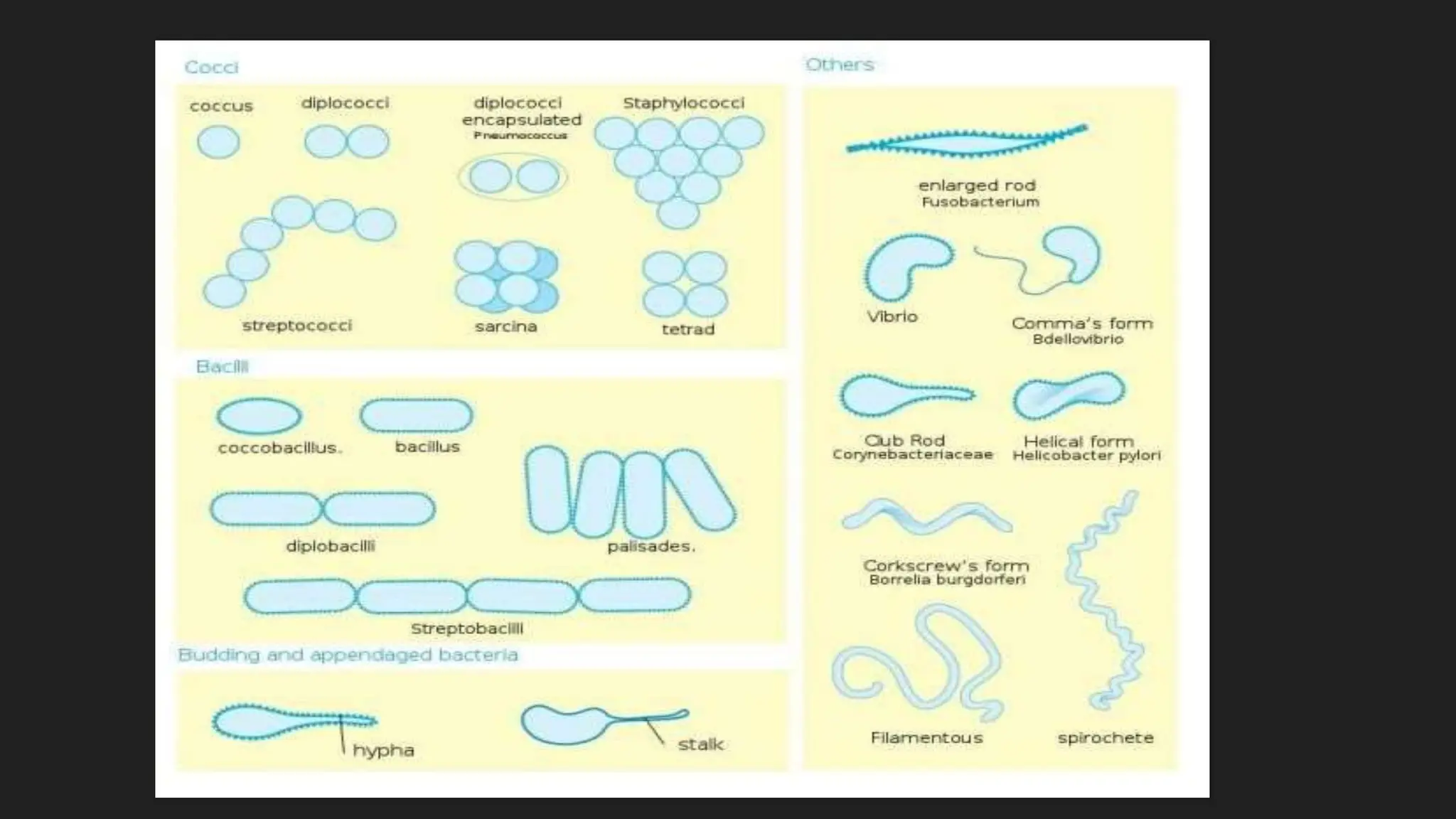
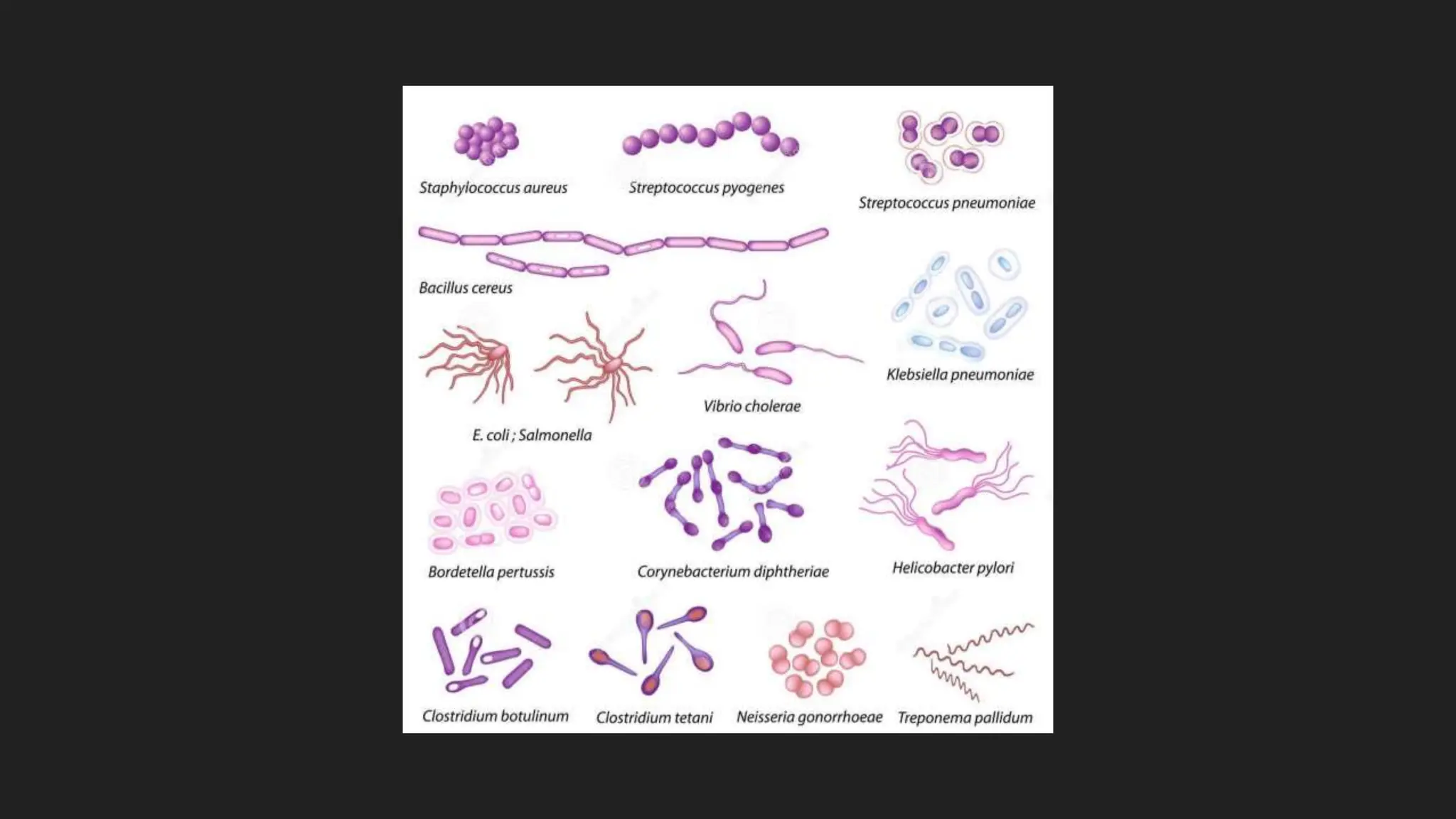
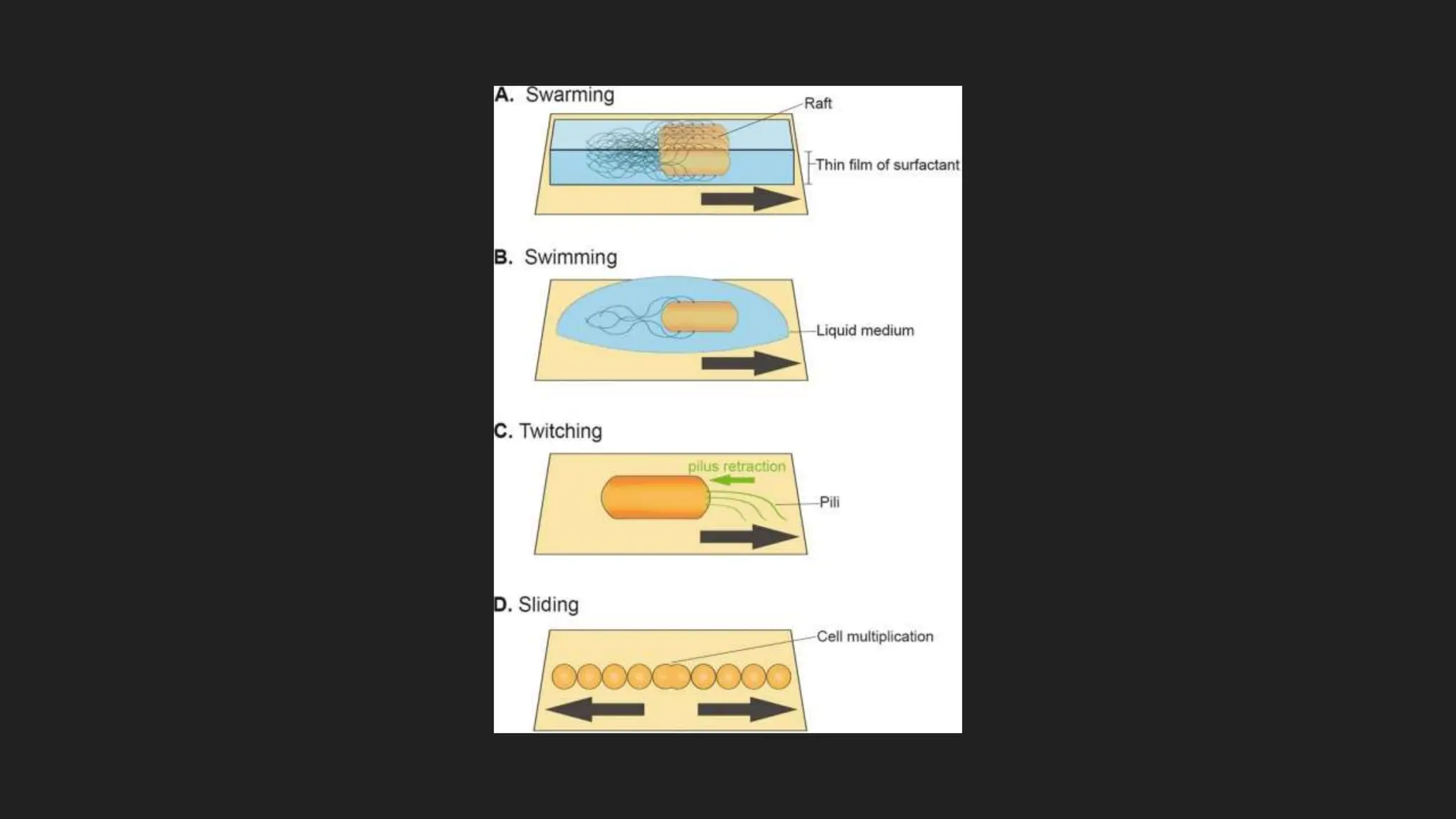
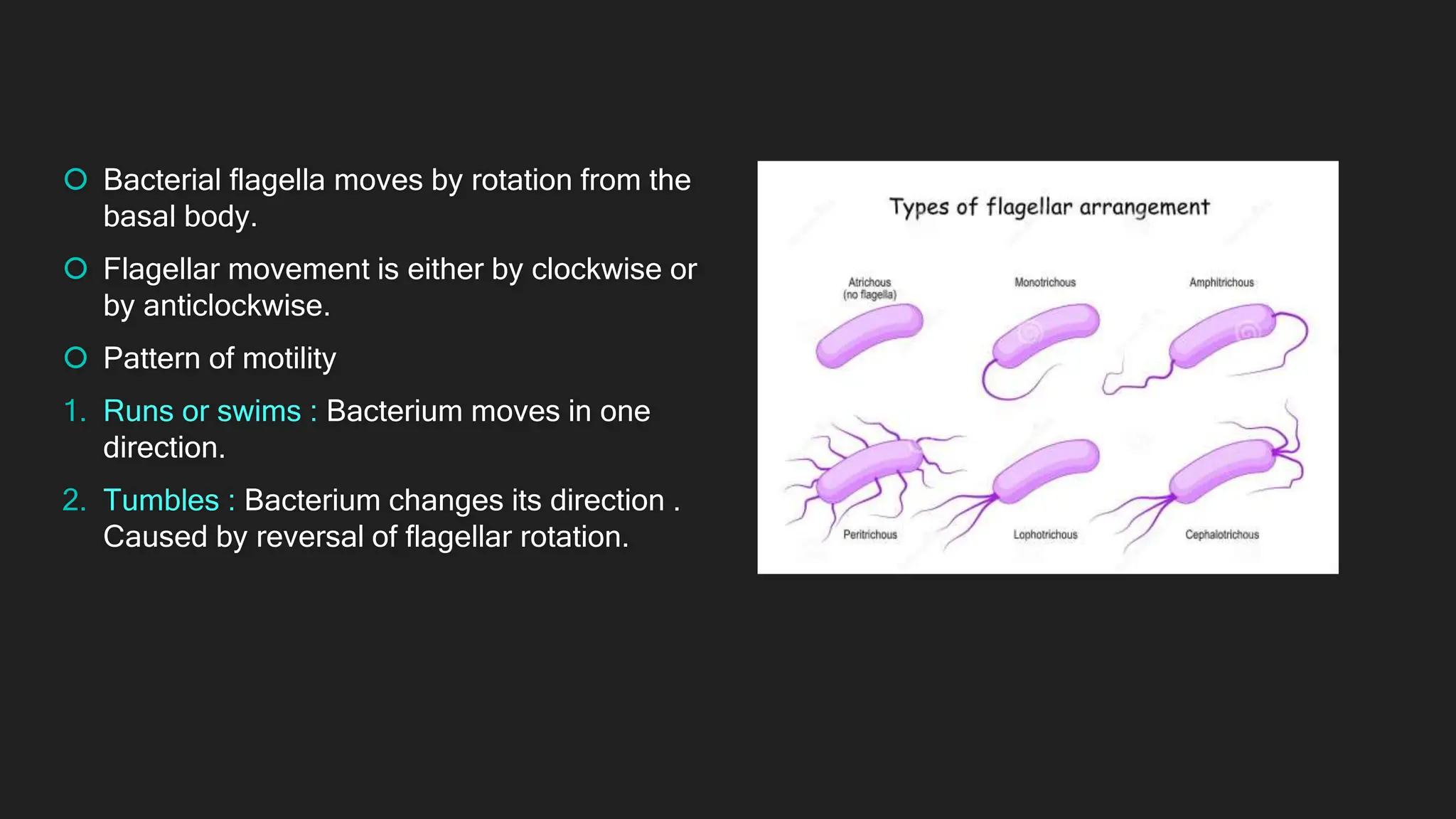
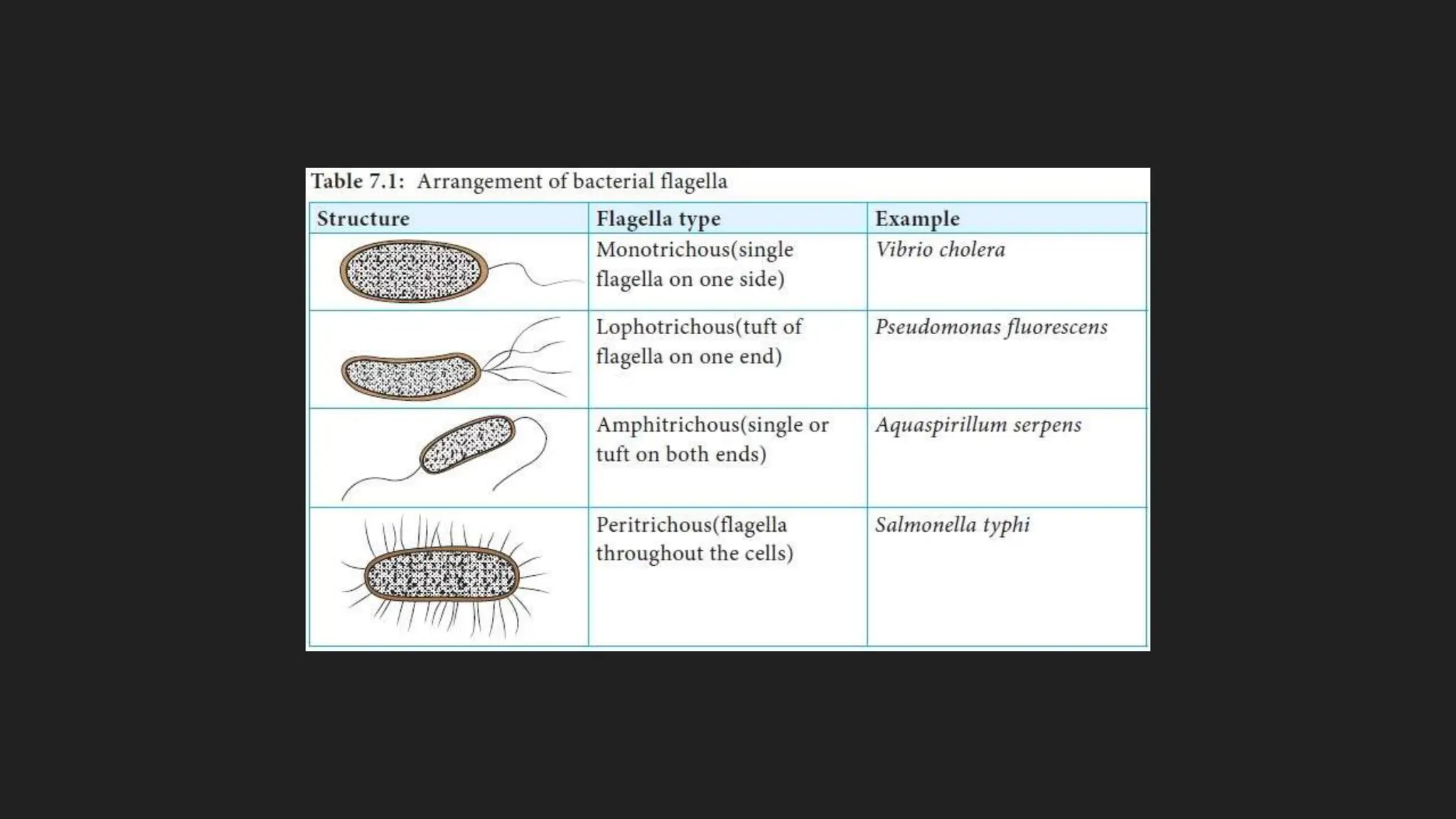
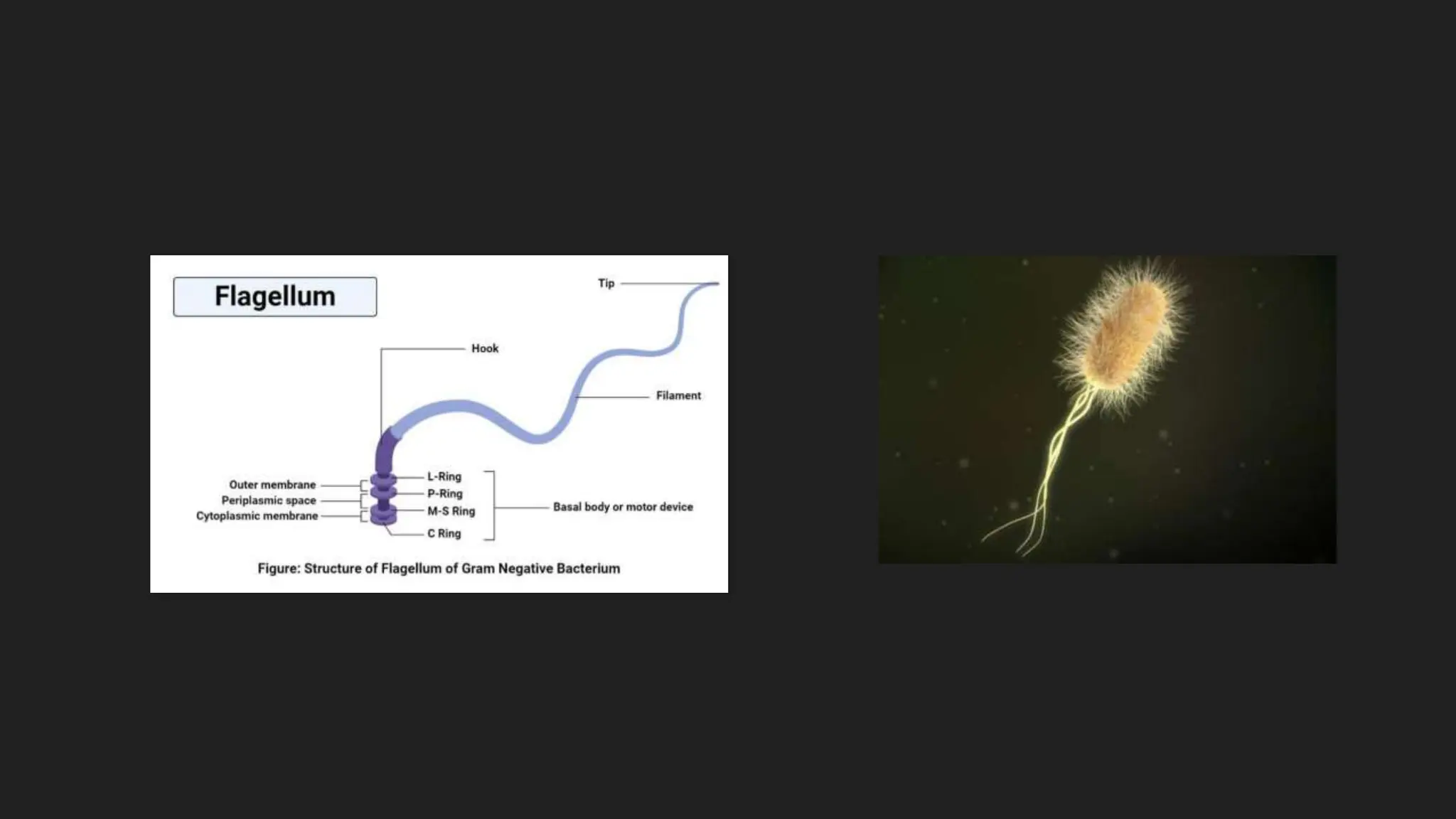
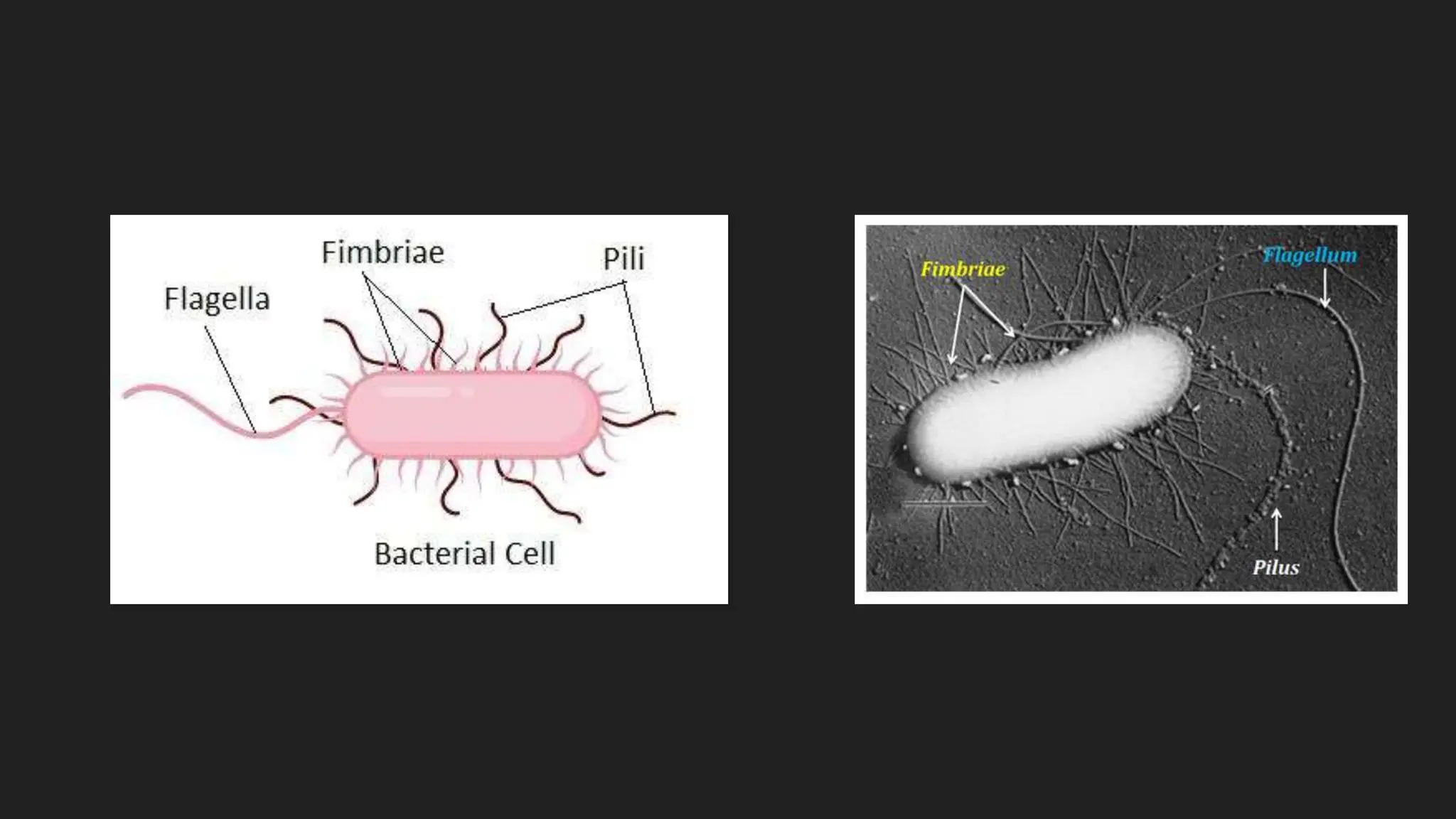
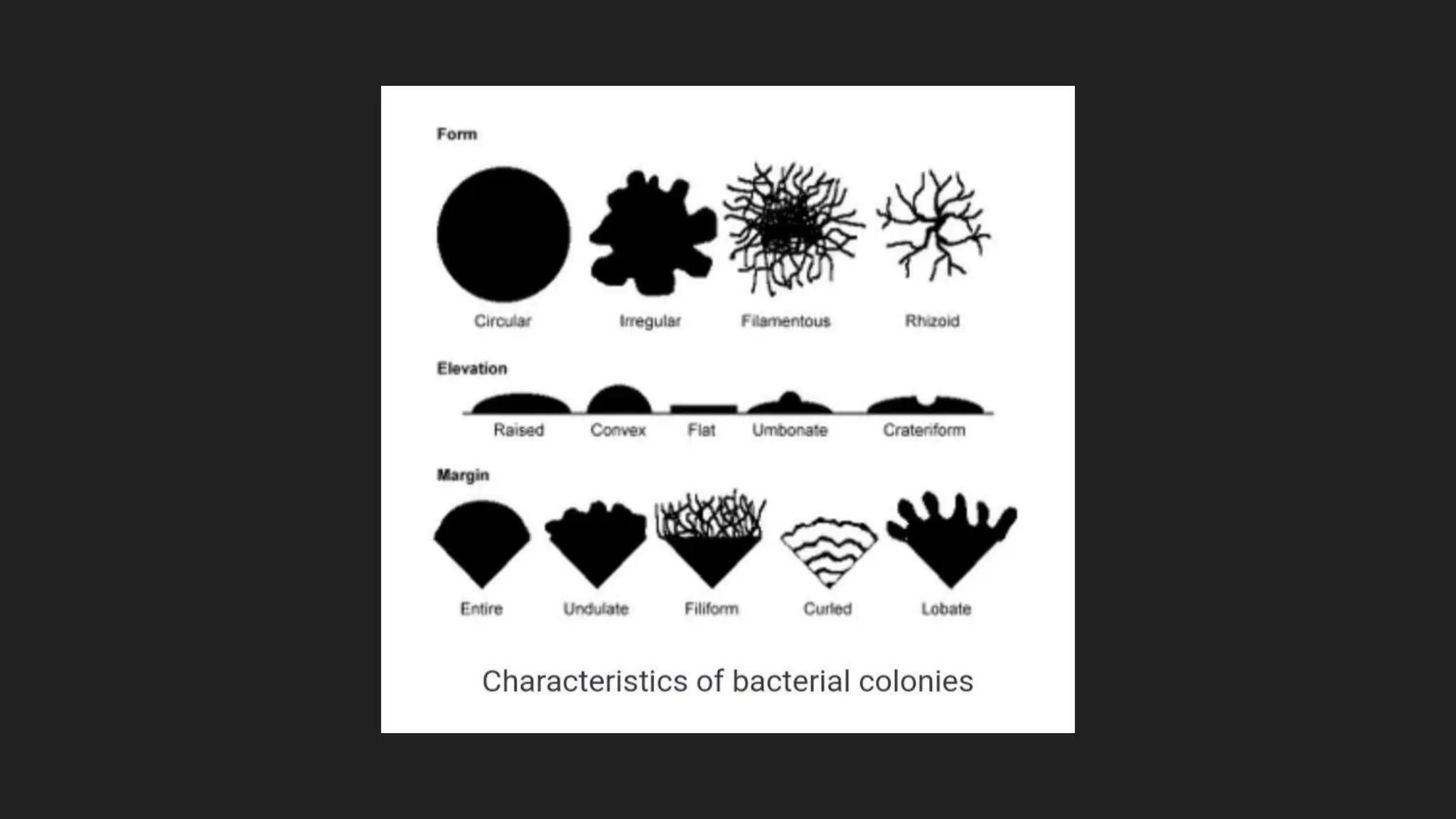
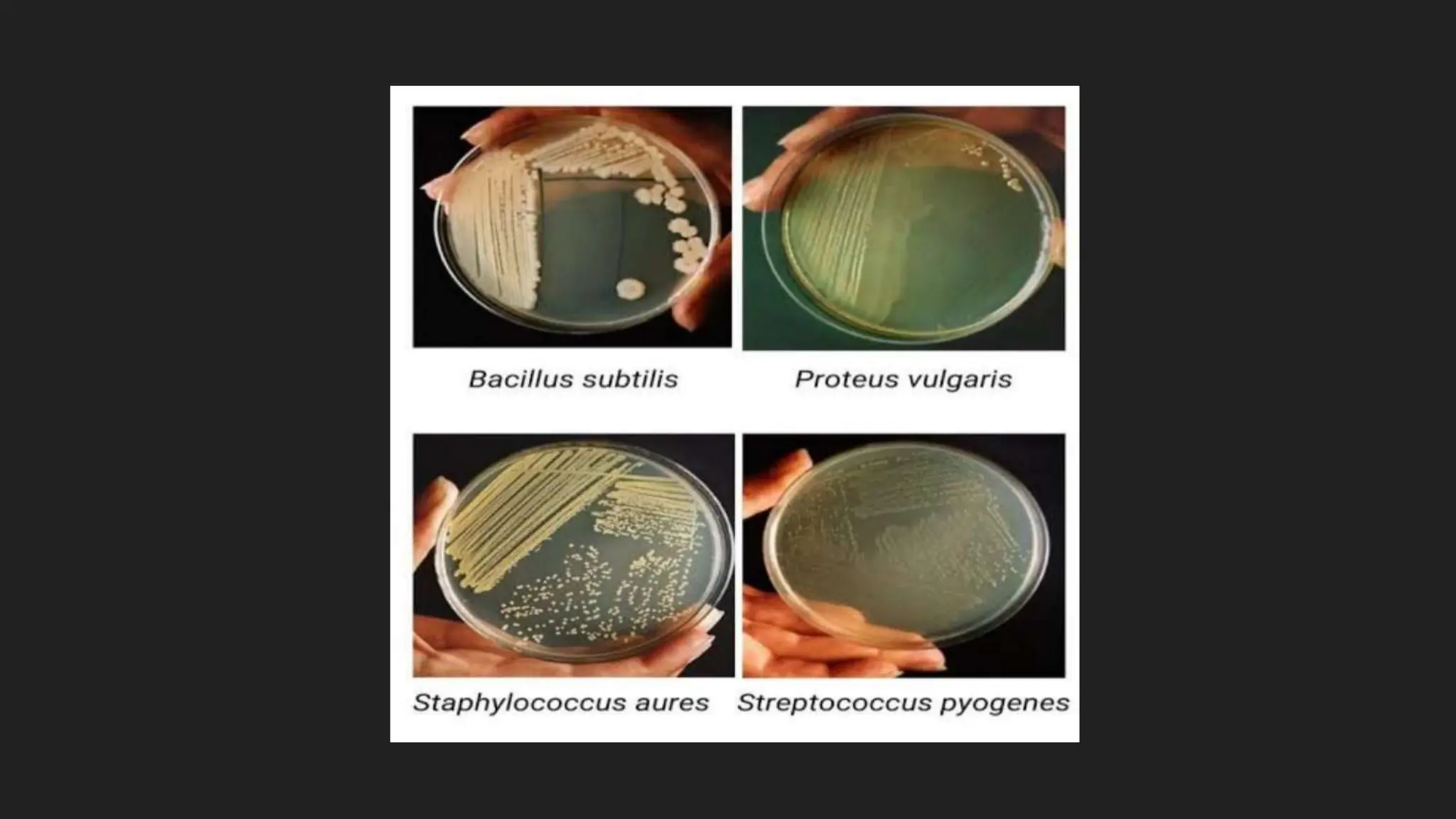
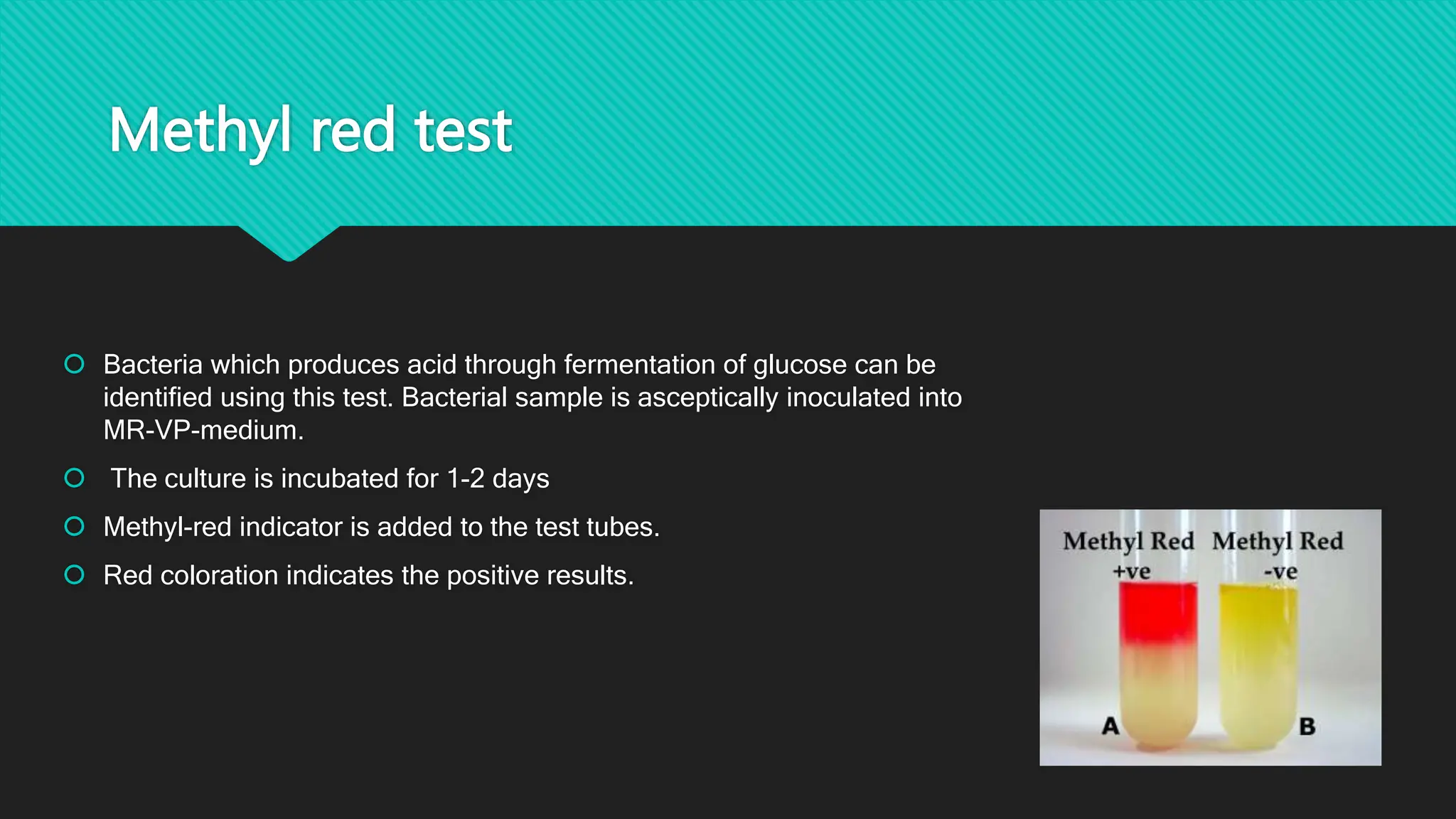
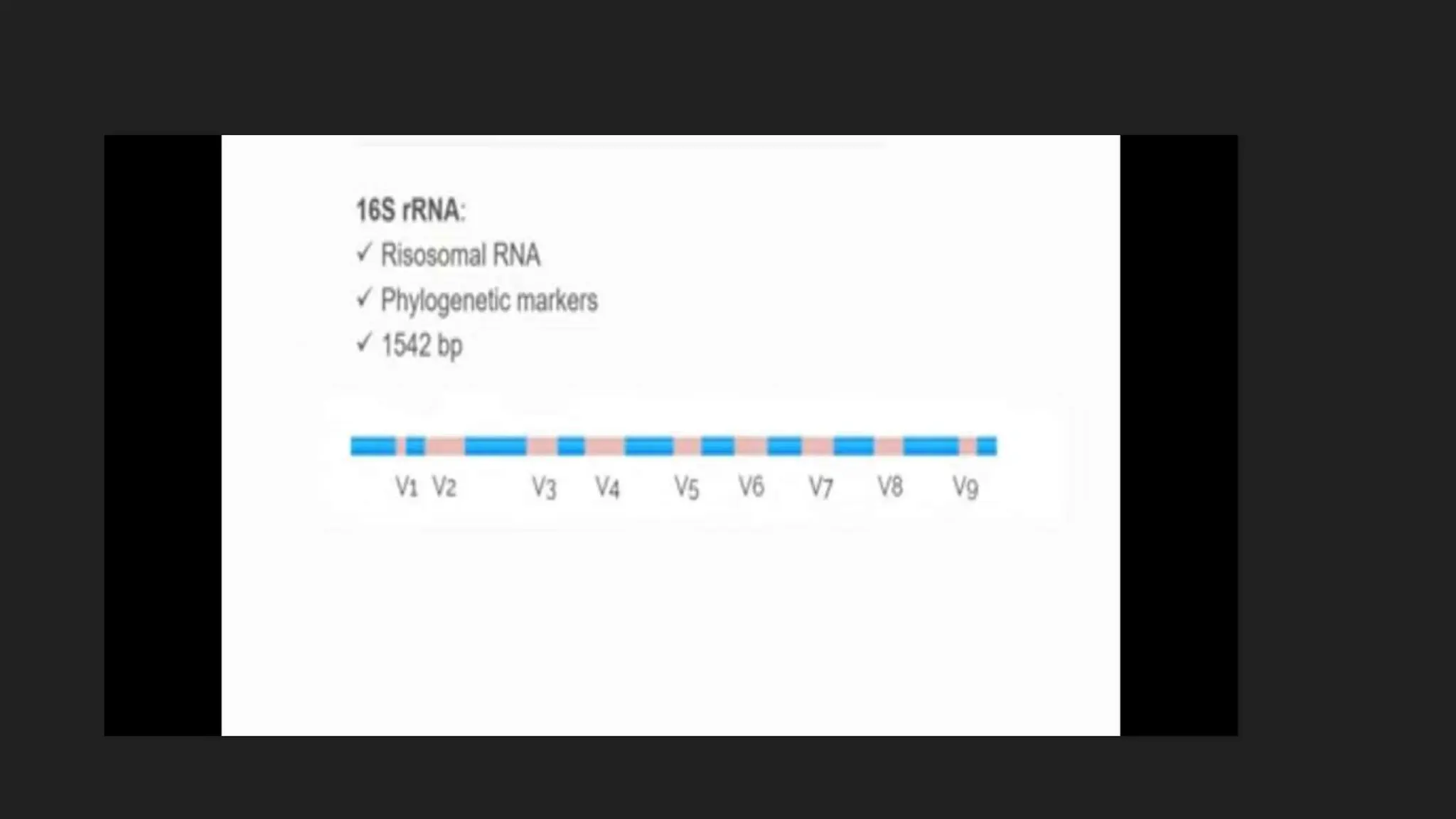
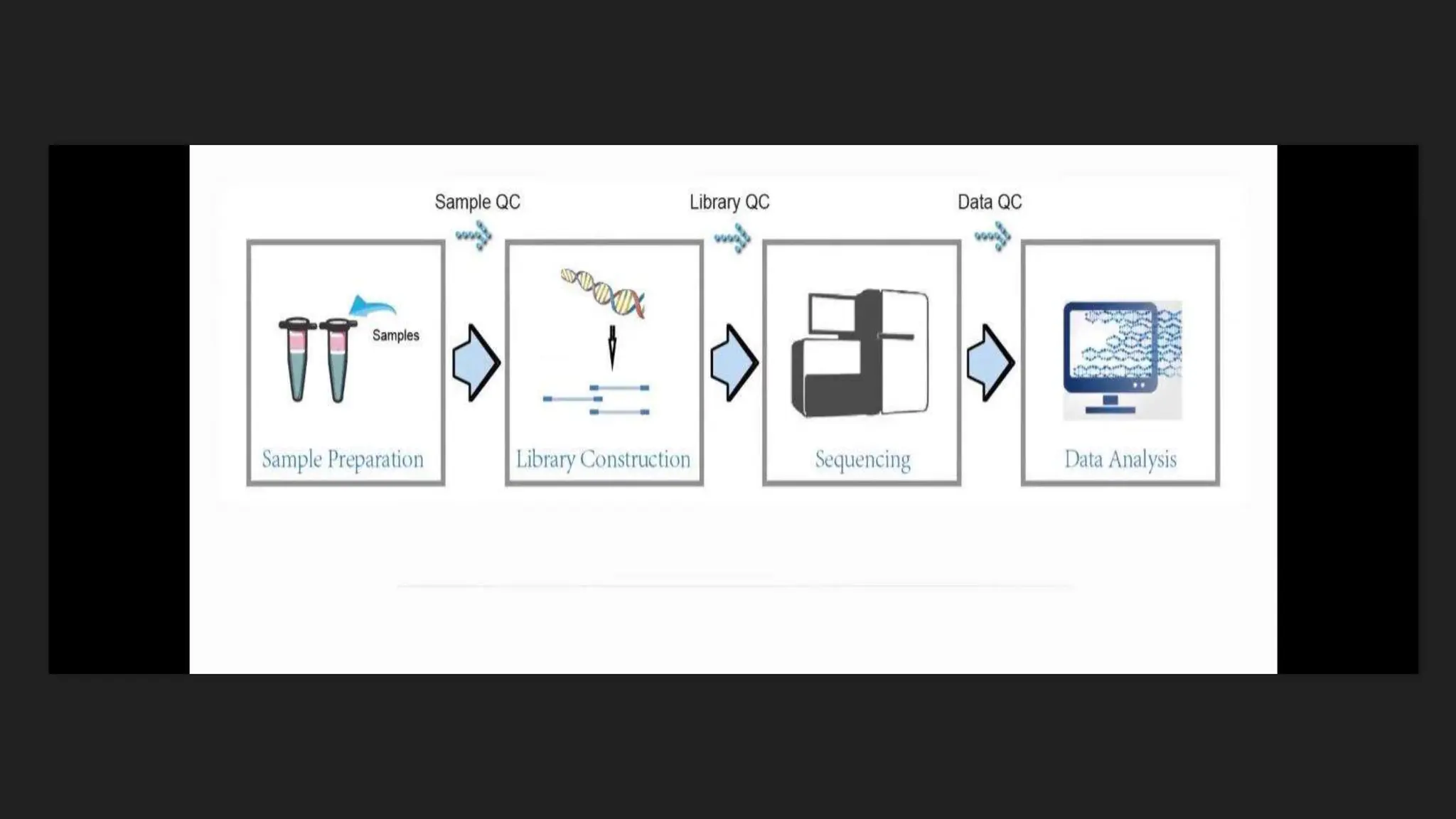
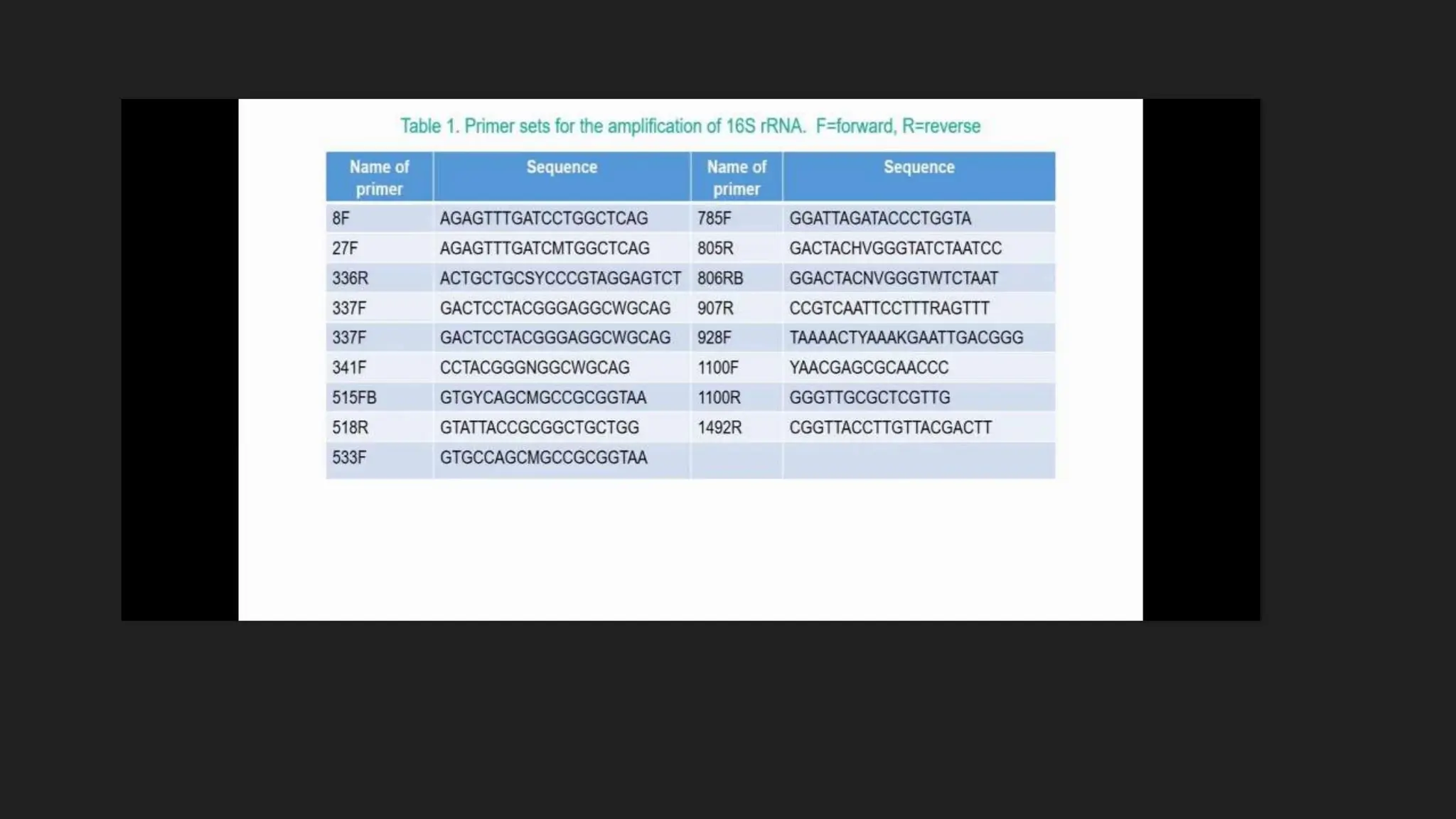
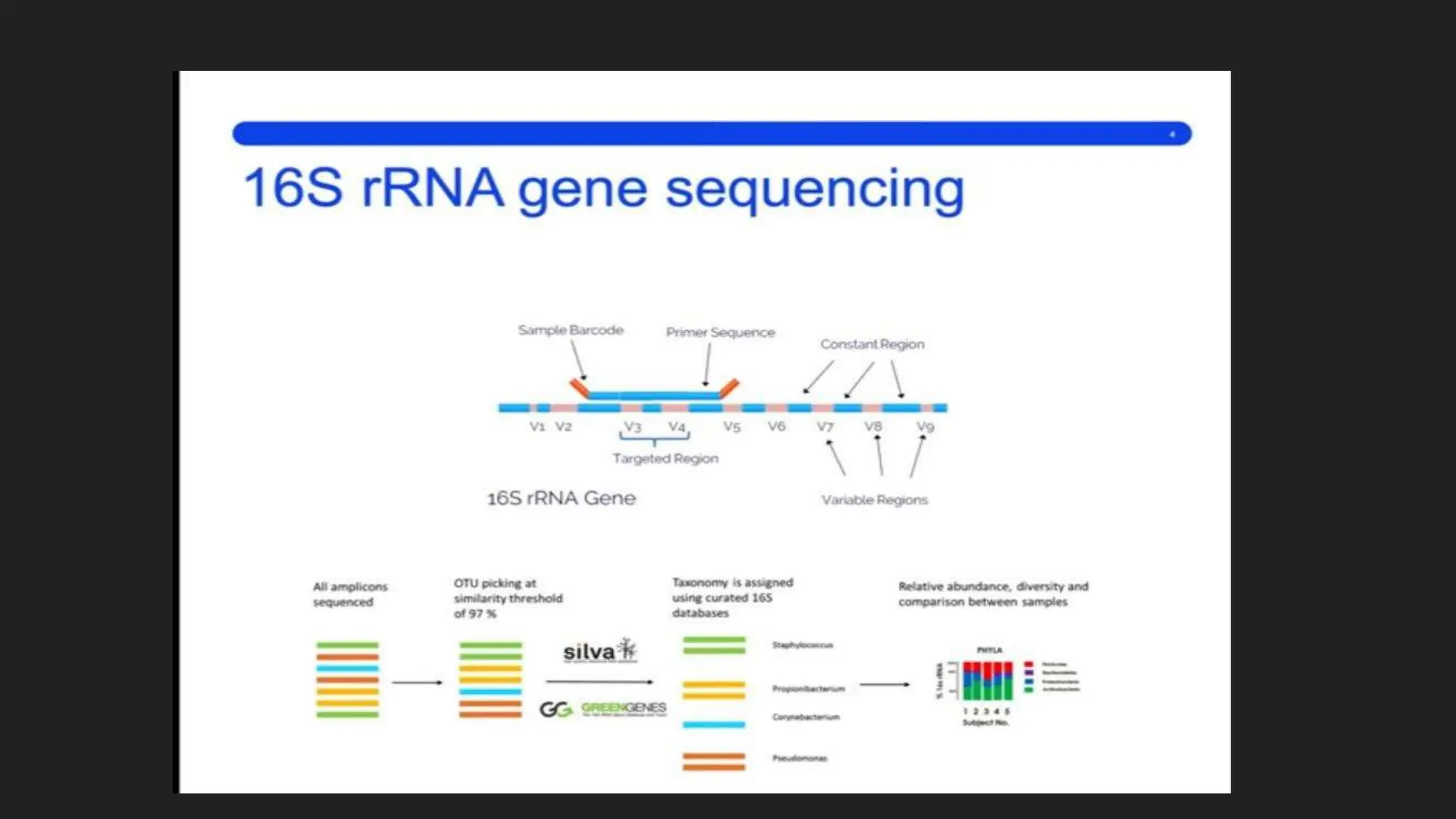
The document discusses various techniques for identifying bacteria, emphasizing morphological and biochemical methods. It explores the classification of bacteria based on their shapes, such as cocci, bacilli, vibrios, spirilla, and spirochetes, and details their movement mechanisms including flagellar, gliding, and spirochaetal movement. Additionally, it covers biochemical tests, specifically the IMViC series, and highlights the importance of 16S rRNA sequencing for bacterial identification.