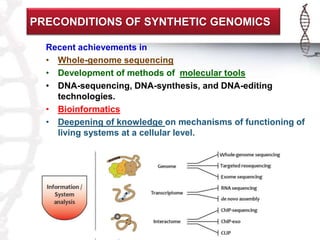
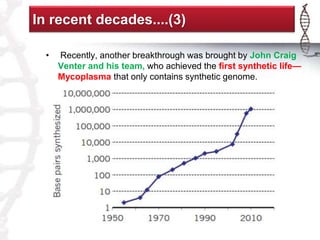
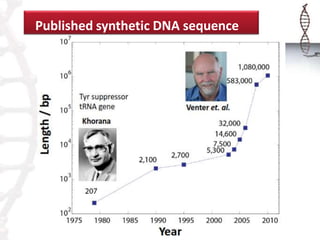
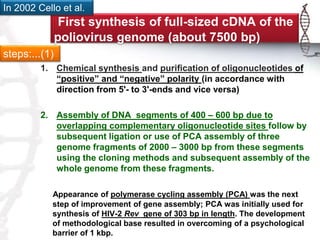
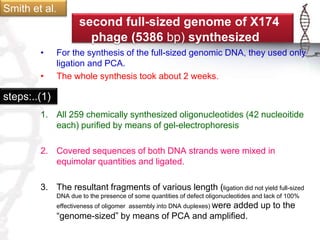
The seminar on synthetic genomes discussed the role of genomic advances in addressing global challenges such as resource scarcity. It highlighted significant achievements in synthetic biology, including the artificial synthesis of genomes and advancements in DNA sequencing and editing technologies. The ultimate goal is to create novel biological systems for various applications, while also noting potential risks of misuse.