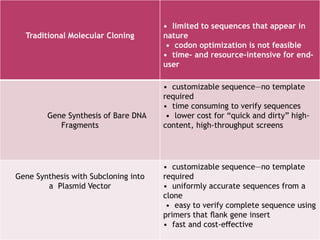
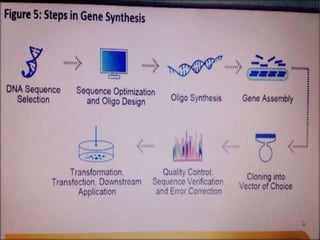
Gene synthesis allows researchers to chemically synthesize DNA sequences without a template. This involves dividing a desired sequence into short oligonucleotides that are synthesized separately and then assembled. Gene synthesis provides advantages over traditional cloning by allowing customizable sequences that do not exist in nature. It has accelerated research in fields like synthetic biology by providing tools that are faster and cheaper to produce than through cloning.