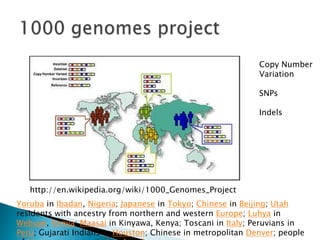
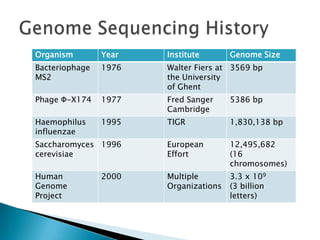
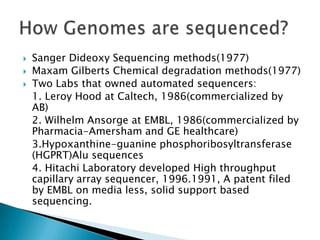
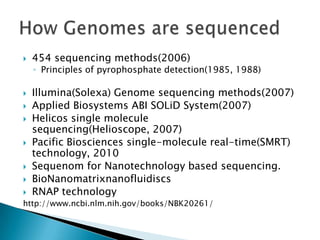
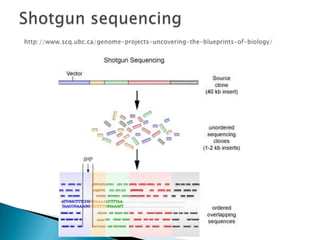
This document provides an overview of genome sequencing. It discusses the history of genome sequencing, from early sequencing of small viruses in the 1970s to larger genomes like yeast and the human genome. The document outlines different sequencing technologies over time, from Sanger sequencing to newer single-molecule approaches. It also summarizes key genome projects like ENCODE and 1000 Genomes that have provided insights into non-coding regulatory elements and human genetic variation.


![ Eukaryotes [2231]
◦ Animal
◦ Fungi
◦ Plants
◦ Protists
◦ Others
Prokaryotes [14268]
Viruses [3219]
Ref:
http://www.ncbi.nlm.nih.gov/genome/brows
e/](https://image.slidesharecdn.com/genomesequencingprojects-120926235720-phpapp02/85/Genome-sequencingprojects-6-320.jpg)

![ JGI – IMG [http://img.jgi.doe.gov/]
Broad
TIGR
WashU
VBI at Virginia Tech](https://image.slidesharecdn.com/genomesequencingprojects-120926235720-phpapp02/85/Genome-sequencingprojects-14-320.jpg)




![Key Findings:
• 80% of the human genome is active!!
– 70,000 promoters and 400,000 enhancers
• 75% of the genome transcribed in some tissue
or other during life time.
• Environment plays great role in switching on
or off of a lot many genes. [Epigenetics]
• Most of the diseases don’t lie with the genes
but the switches!!
• Dark matters controlling the genes are
physically close to the genes they control.](https://image.slidesharecdn.com/genomesequencingprojects-120926235720-phpapp02/85/Genome-sequencingprojects-22-320.jpg)