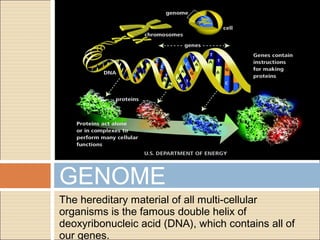
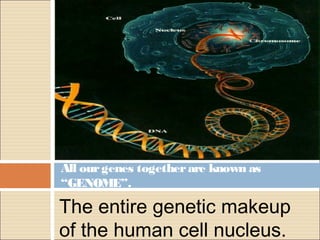
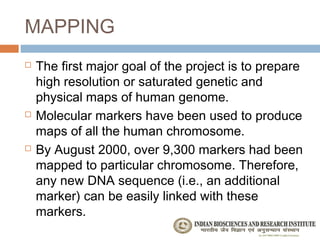
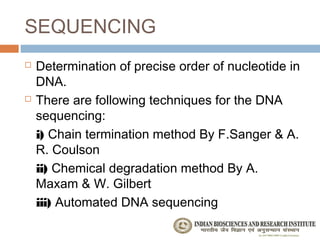
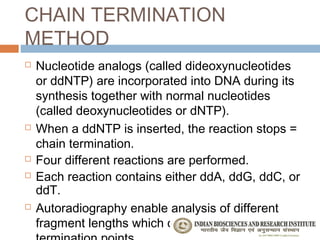
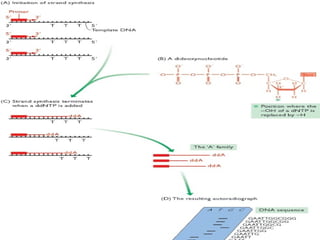
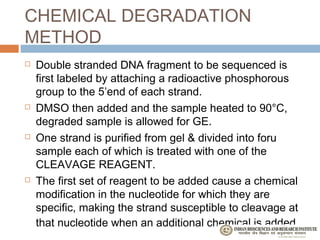
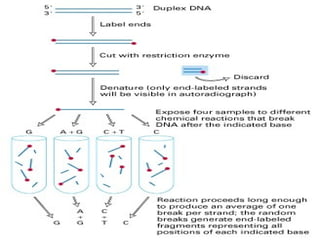
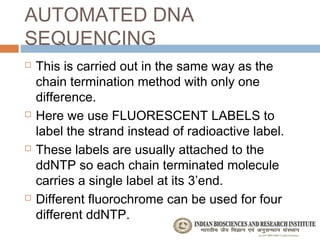
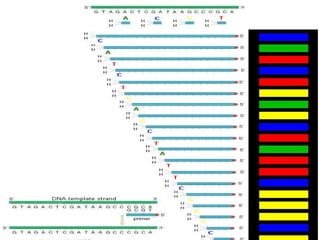
The document provides an overview of the Human Genome Project (HGP). It describes the HGP's goal of mapping and sequencing the entire human genome. The HGP was an international research effort that worked alongside a private company, Celera Genomics, to complete a rough draft of the human genome by 2000. The completion of the HGP marked a major scientific achievement and has transformed fields like medicine, biotechnology, and genetics by providing a comprehensive map of the human genetic code.















![ETHICAL, LEGAL & SOCIAL
ISSUES
[ELSI]](https://image.slidesharecdn.com/humangenomeprojectibri-130109235449-phpapp01/85/Human-genome-project-ibri-27-320.jpg)






![BIOTECHNOLOGY
Production of useful protein products for use
in medicine, agriculture, bioremediation and
pharmaceutical industries.
Antibiotics
Protein replacement (factor VIII, TPA,
streptokinase, insulin, interferon…)
BT insecticide toxin (from Ba c illus
thuring ie ns is )
Herbicide resistance (glyphosate resistance)
Bioengineered foods [e.g. Flavr Savr tomato
(antisense – polygalacturonase) to delay
rotting]](https://image.slidesharecdn.com/humangenomeprojectibri-130109235449-phpapp01/85/Human-genome-project-ibri-34-320.jpg)