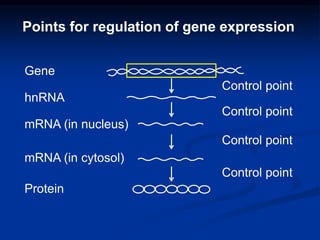
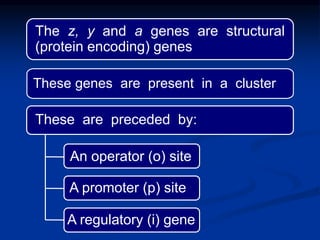
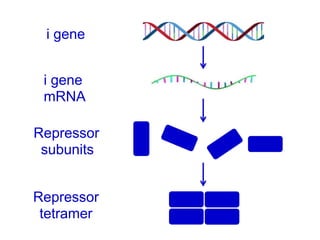
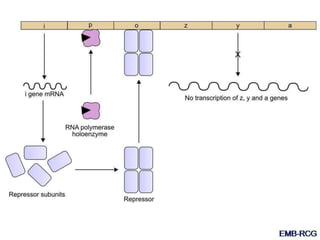
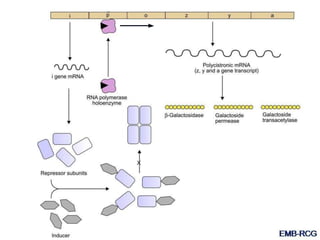
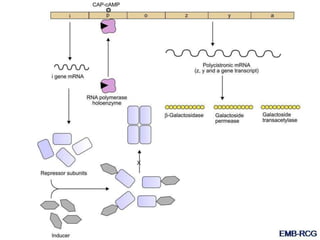
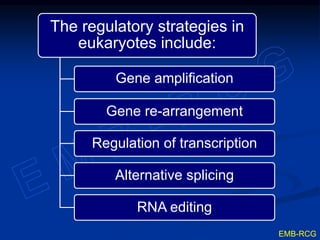
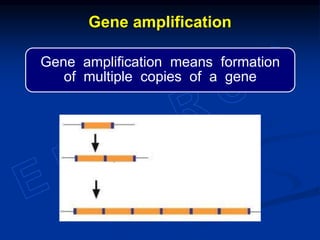
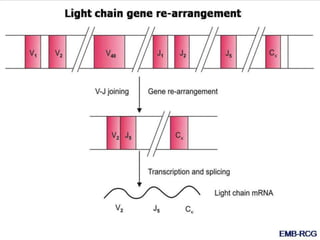
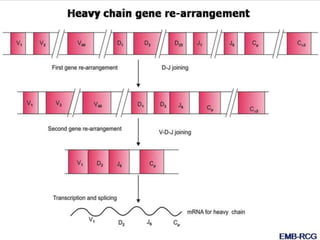
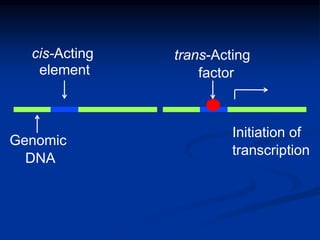
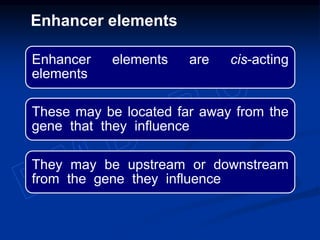
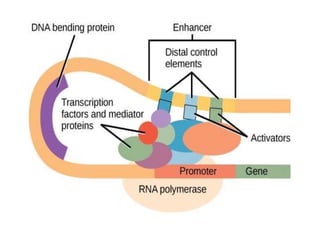
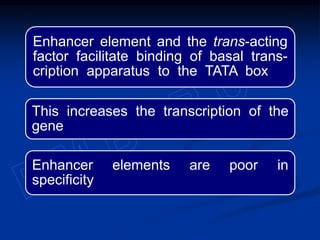
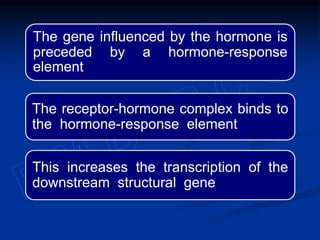
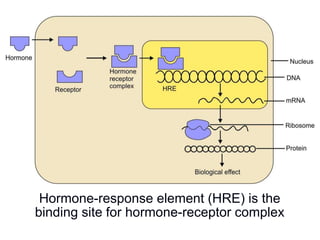
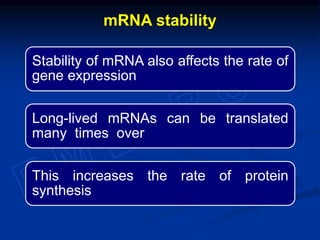
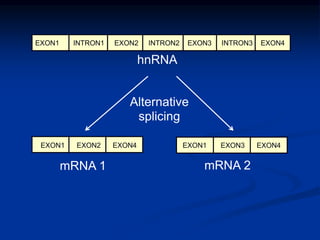
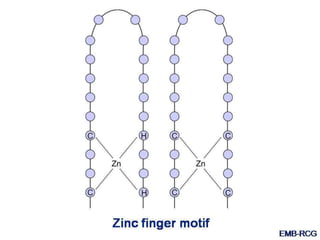
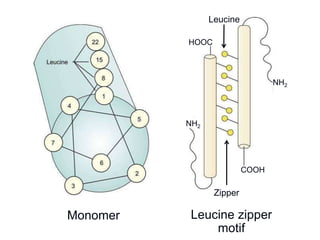
The document discusses the regulation of gene expression, detailing mechanisms in both prokaryotes and eukaryotes, including transcription, RNA modification, and translation. Key regulatory components such as operons, enhancers, and silencers are explained, highlighting their roles in gene expression control. It also covers methods like gene amplification and alternative splicing that contribute to protein diversity and functional adaptation in organisms.