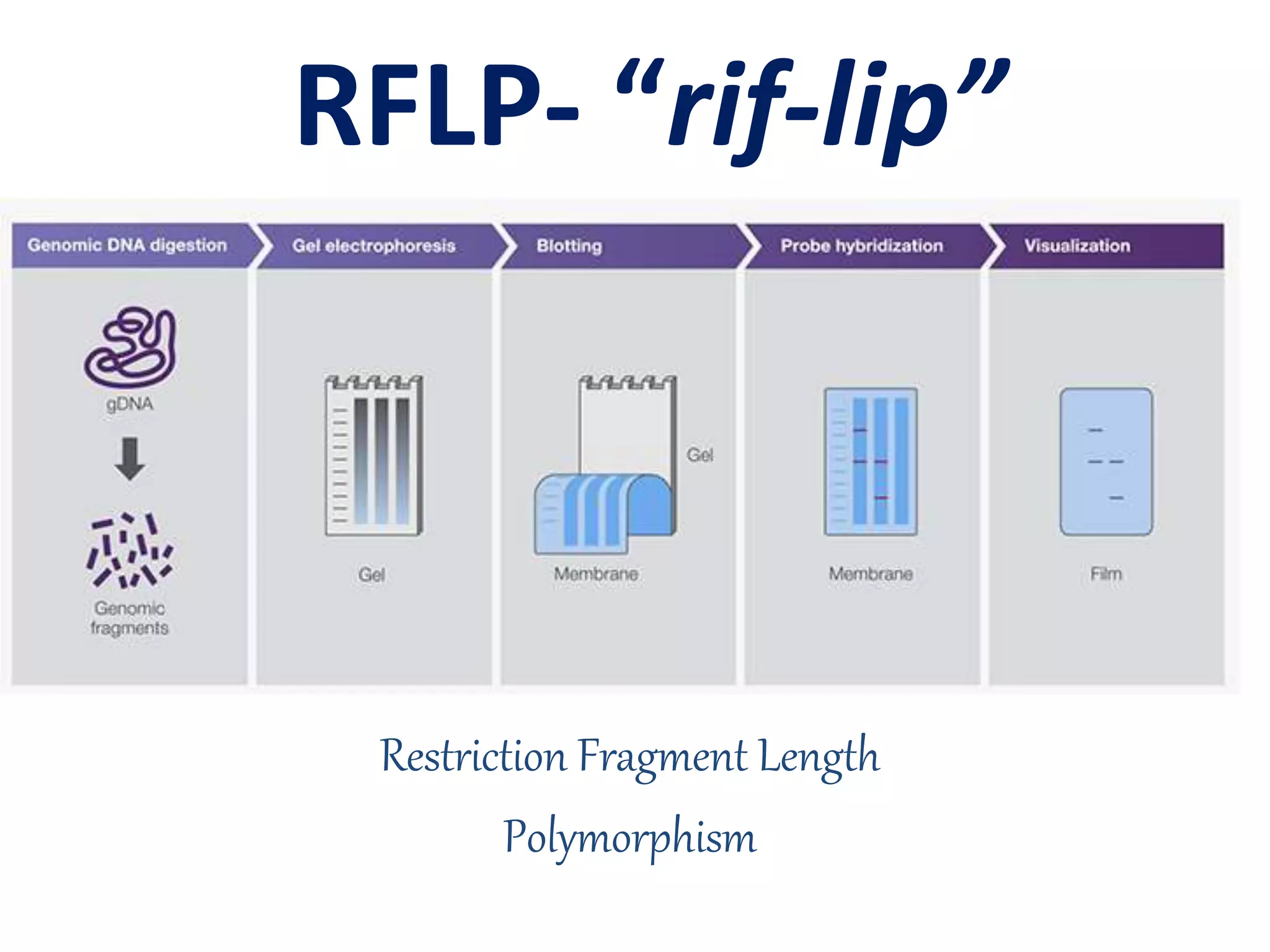
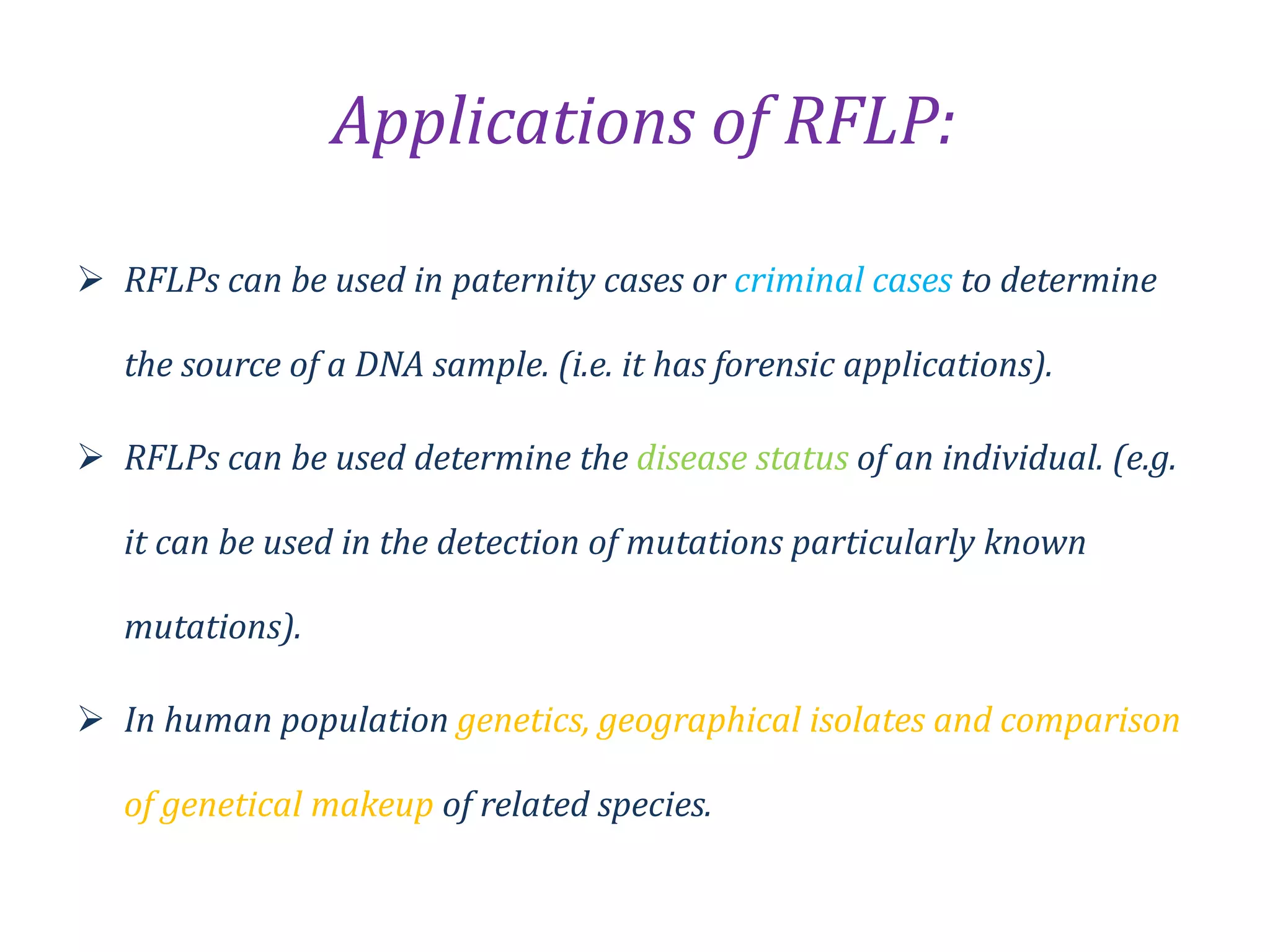
Restriction Fragment Length Polymorphism (RFLP) is a technique for analyzing genetic variation by cutting DNA with restriction enzymes and visualizing differences in fragment lengths, primarily popularized in the 1980s. Technological advancements in restriction enzymes and DNA hybridization enabled the development of this method, which is now extensively used in genetics for applications such as paternity testing and disease mutation detection. However, RFLP requires large amounts of DNA and can be time-consuming, making it less suitable for some modern applications compared to newer methods like PCR.