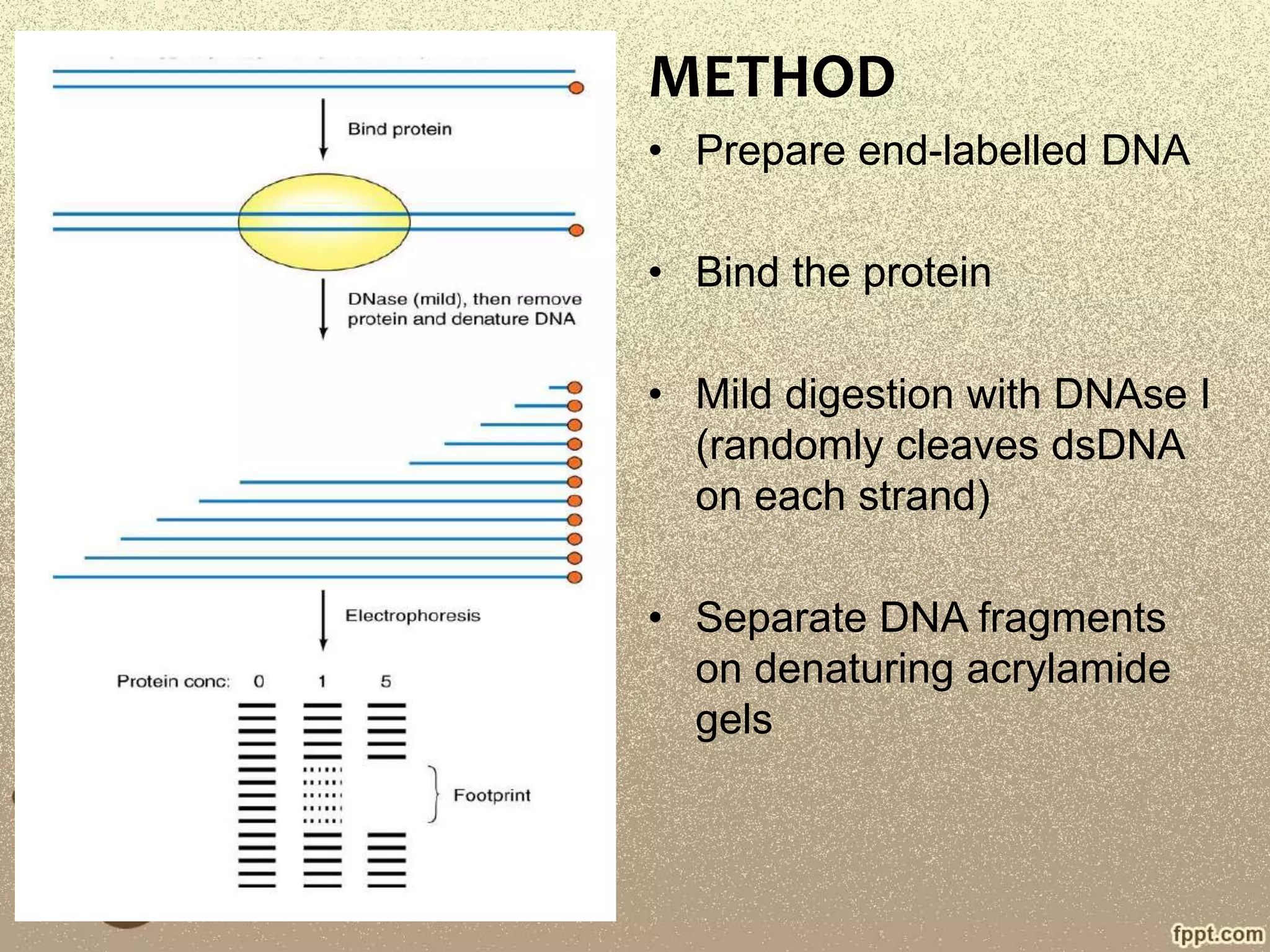
DNA footprinting is a technique used to identify where proteins bind to DNA. It was developed in 1978 and works by treating DNA with enzymes or chemicals that cut DNA, except for regions bound by proteins. This leaves a "footprint" where the protein is bound and protects the DNA. There are two main types: DNase I footprinting cuts DNA randomly except where proteins are bound, while DMS footprinting modifies DNA bases except where proteins protect them from modification. The cut or modified DNA is then run on a gel to identify the protein binding site. DNA footprinting is useful for mapping transcription factor binding sites and studying protein-DNA interactions.