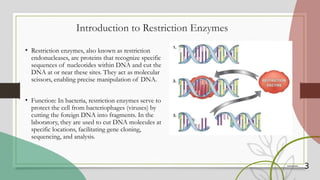
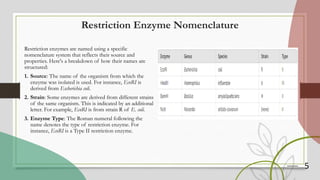
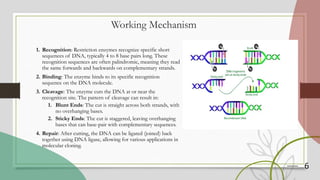
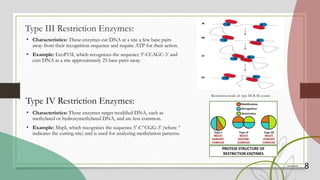
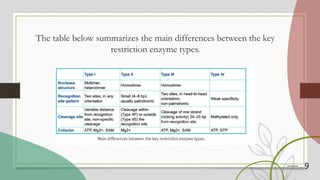
The document discusses restriction enzymes, which are proteins that cut DNA at specific sequences, acting as molecular scissors for gene manipulation. It covers their history, types (I, II, III, and IV), mechanisms of action, and applications in genetic engineering, DNA fingerprinting, gene mapping, and mutation analysis. The conclusion emphasizes the significance of these enzymes in advancing molecular biology and biotechnology.