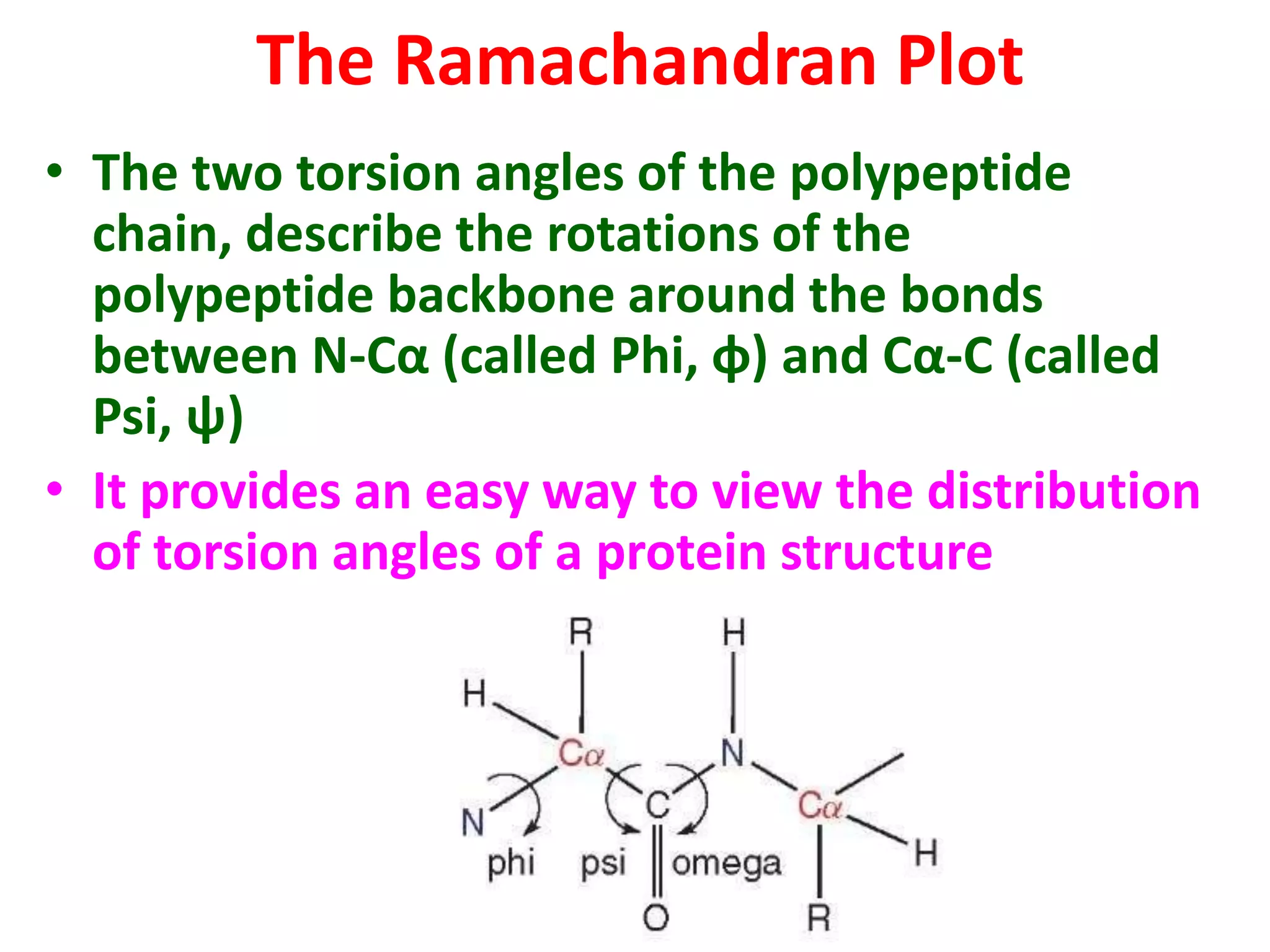
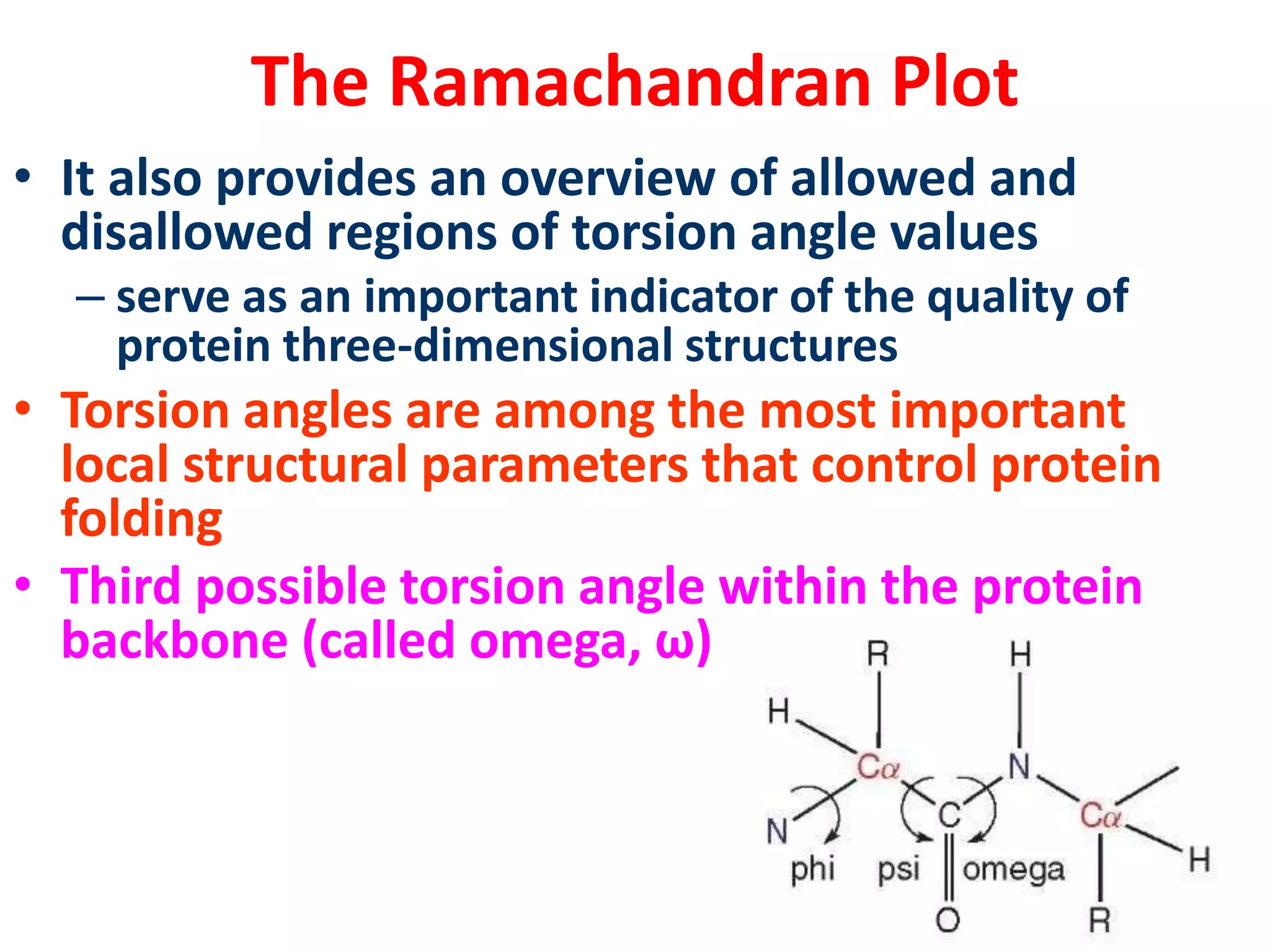
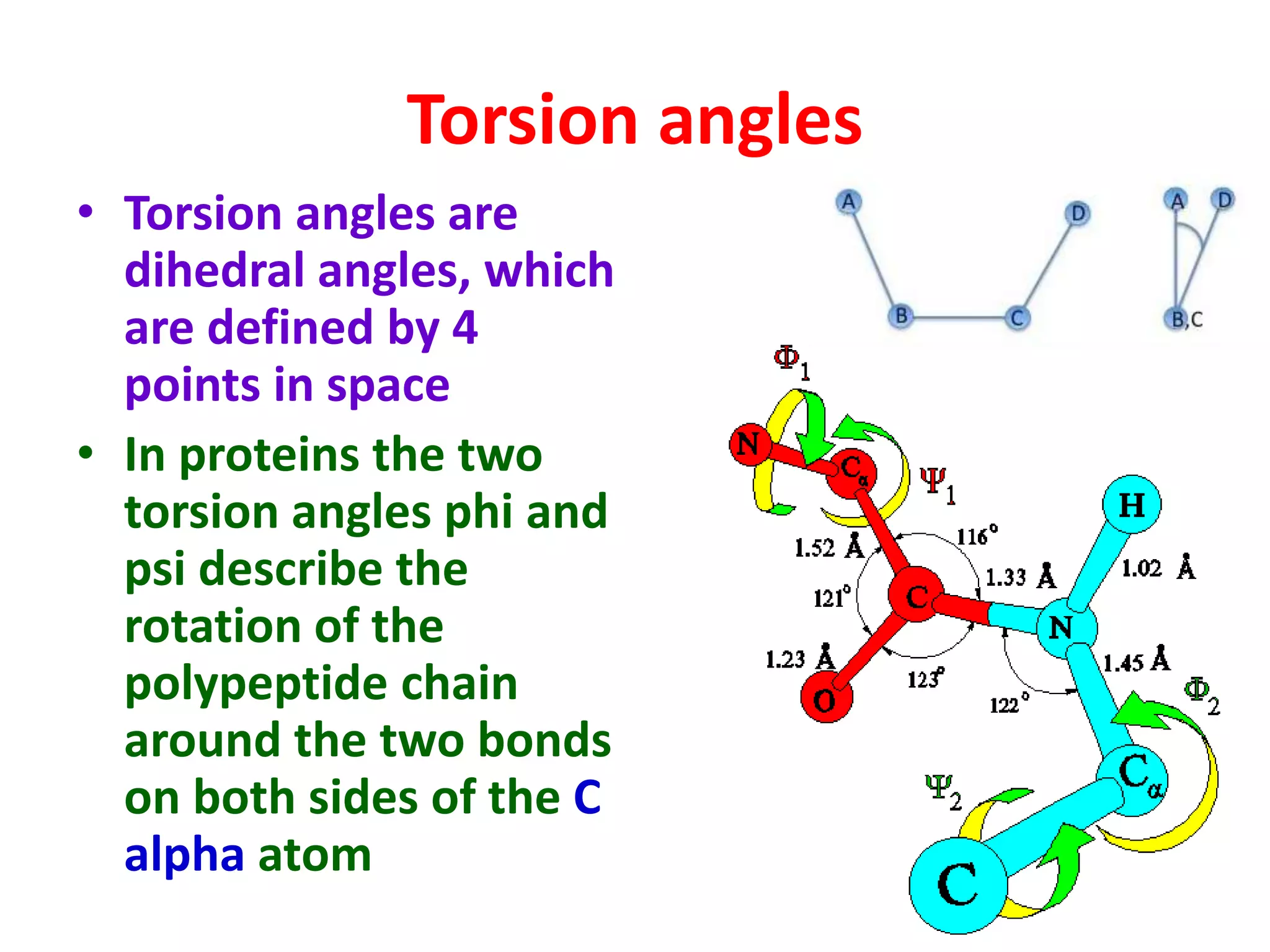
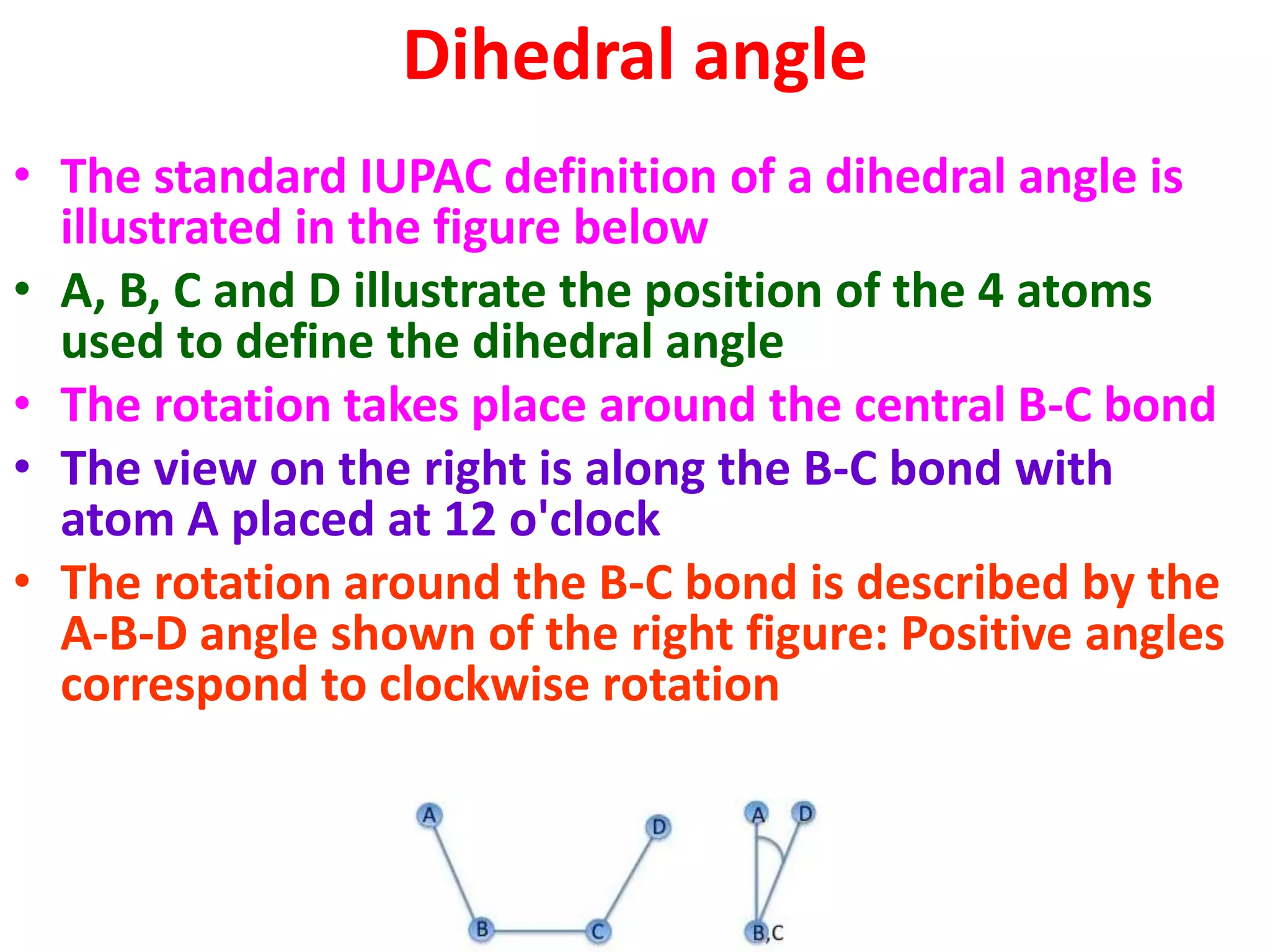
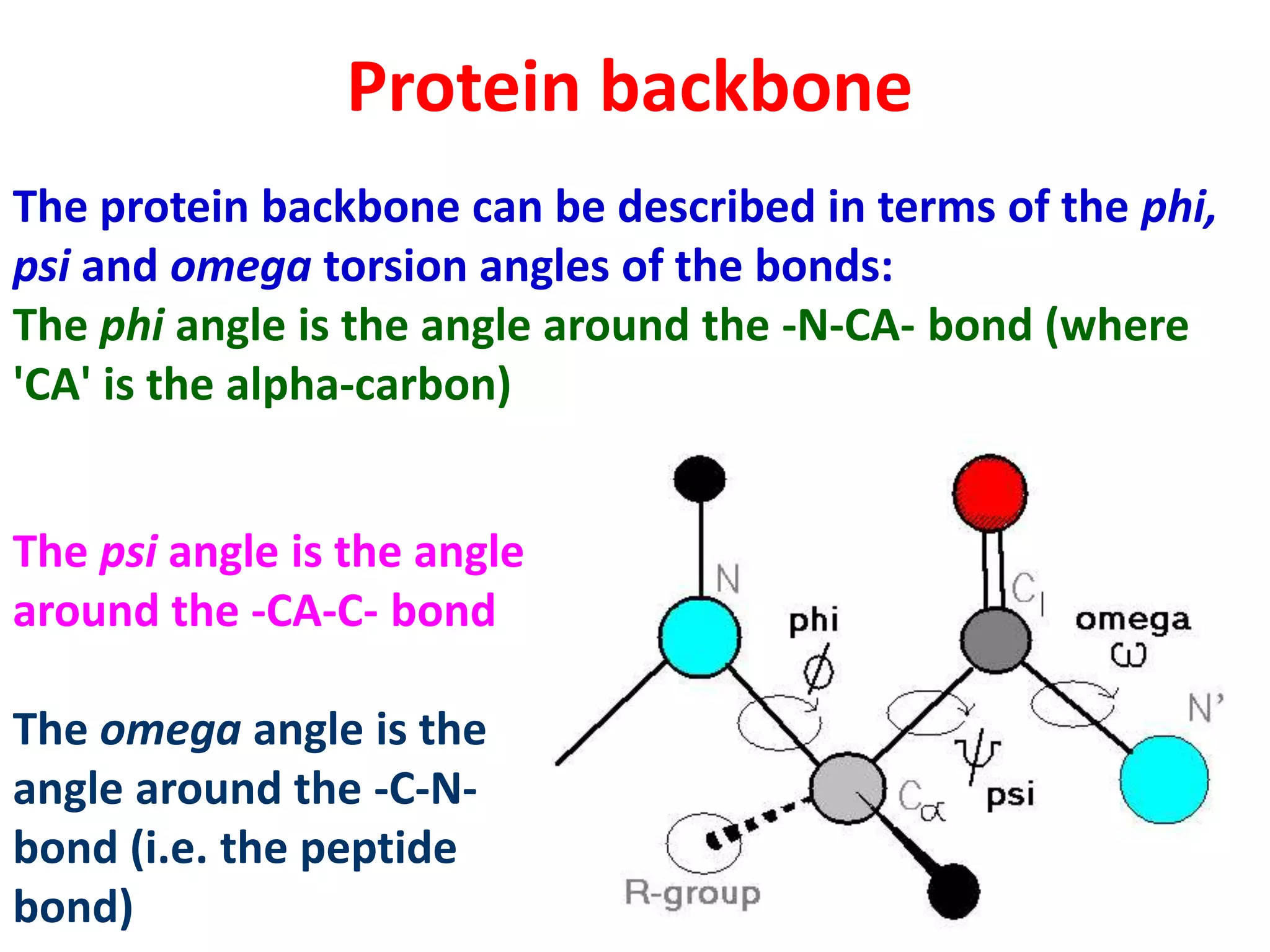
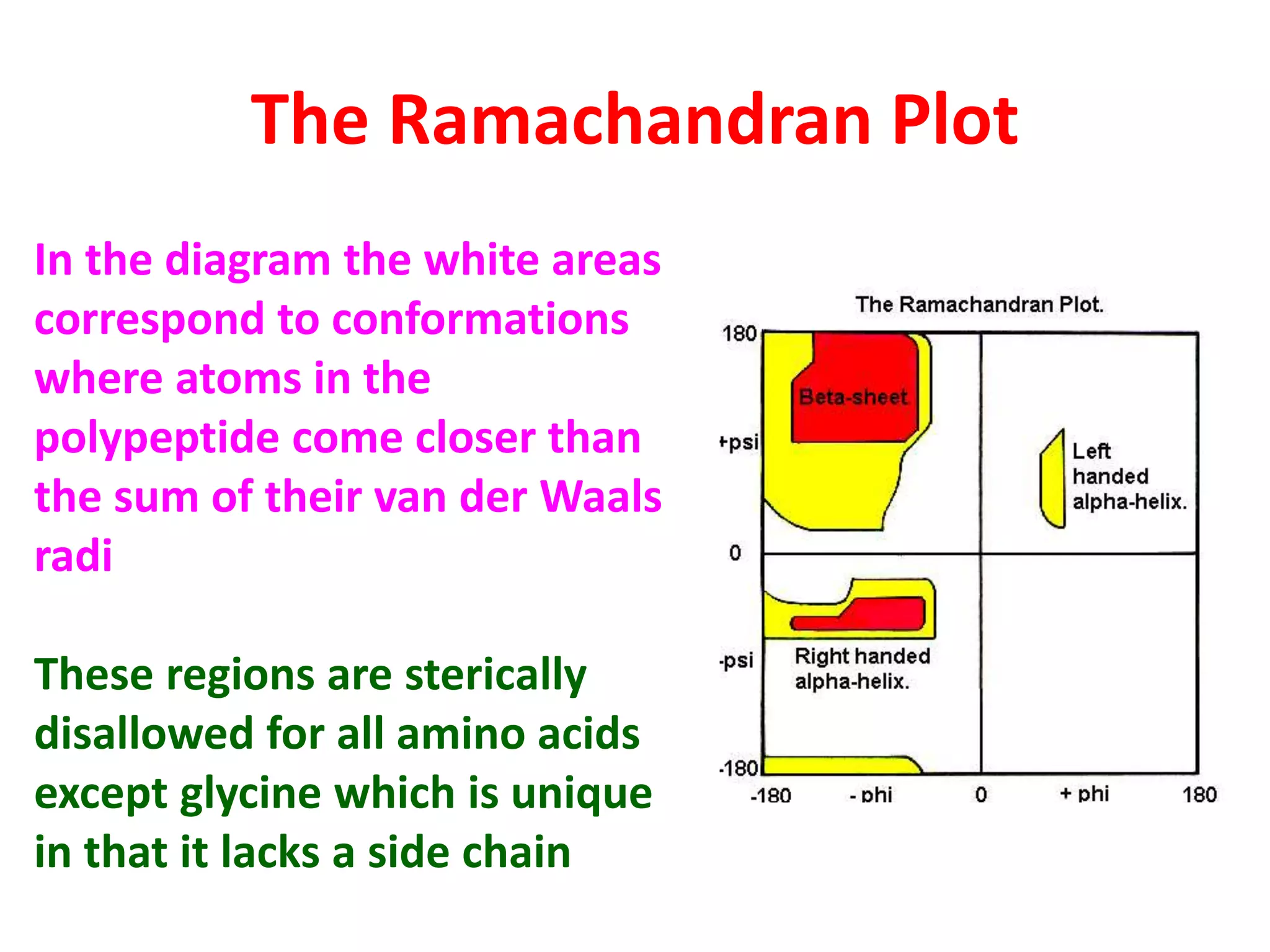
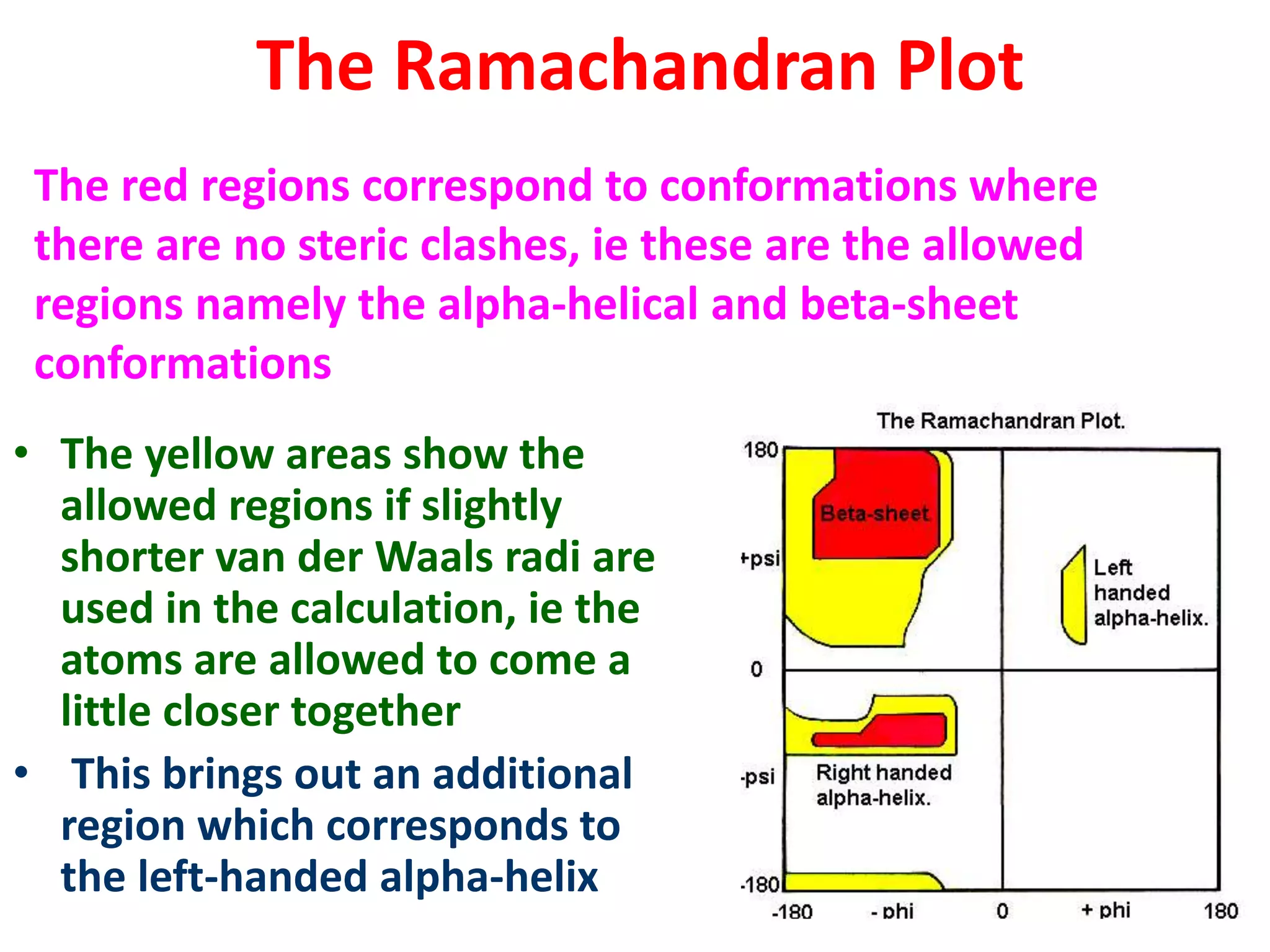
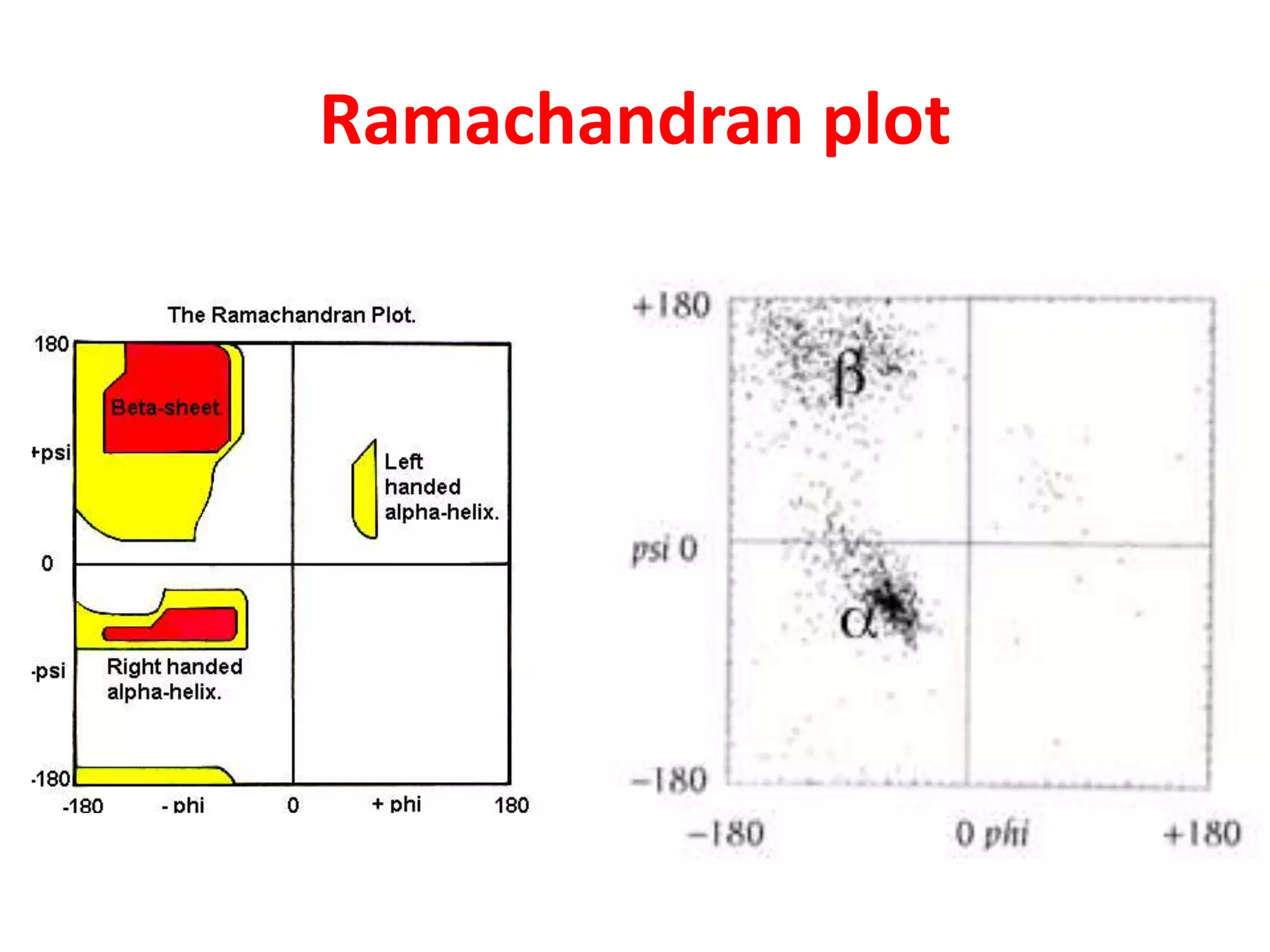
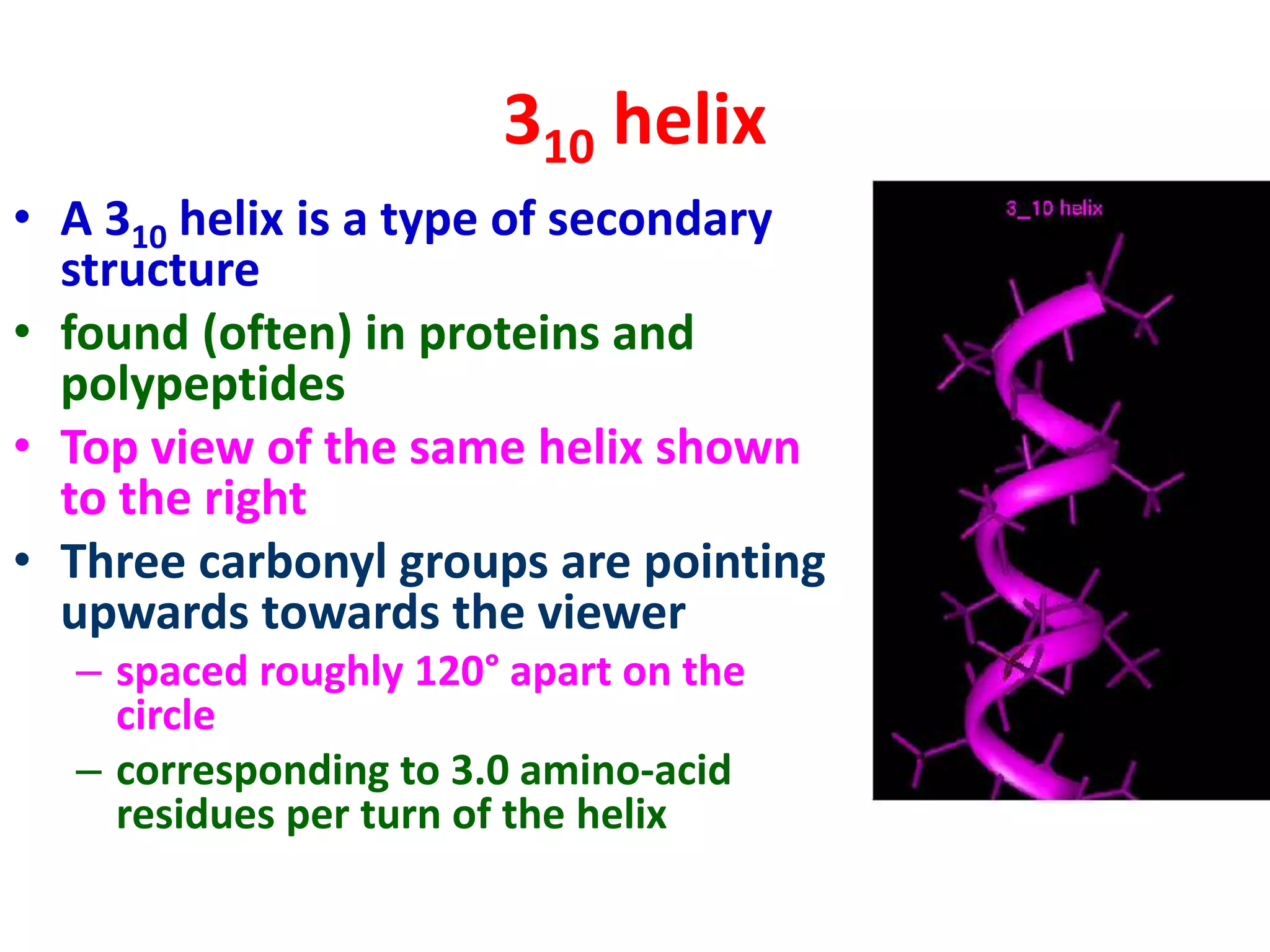
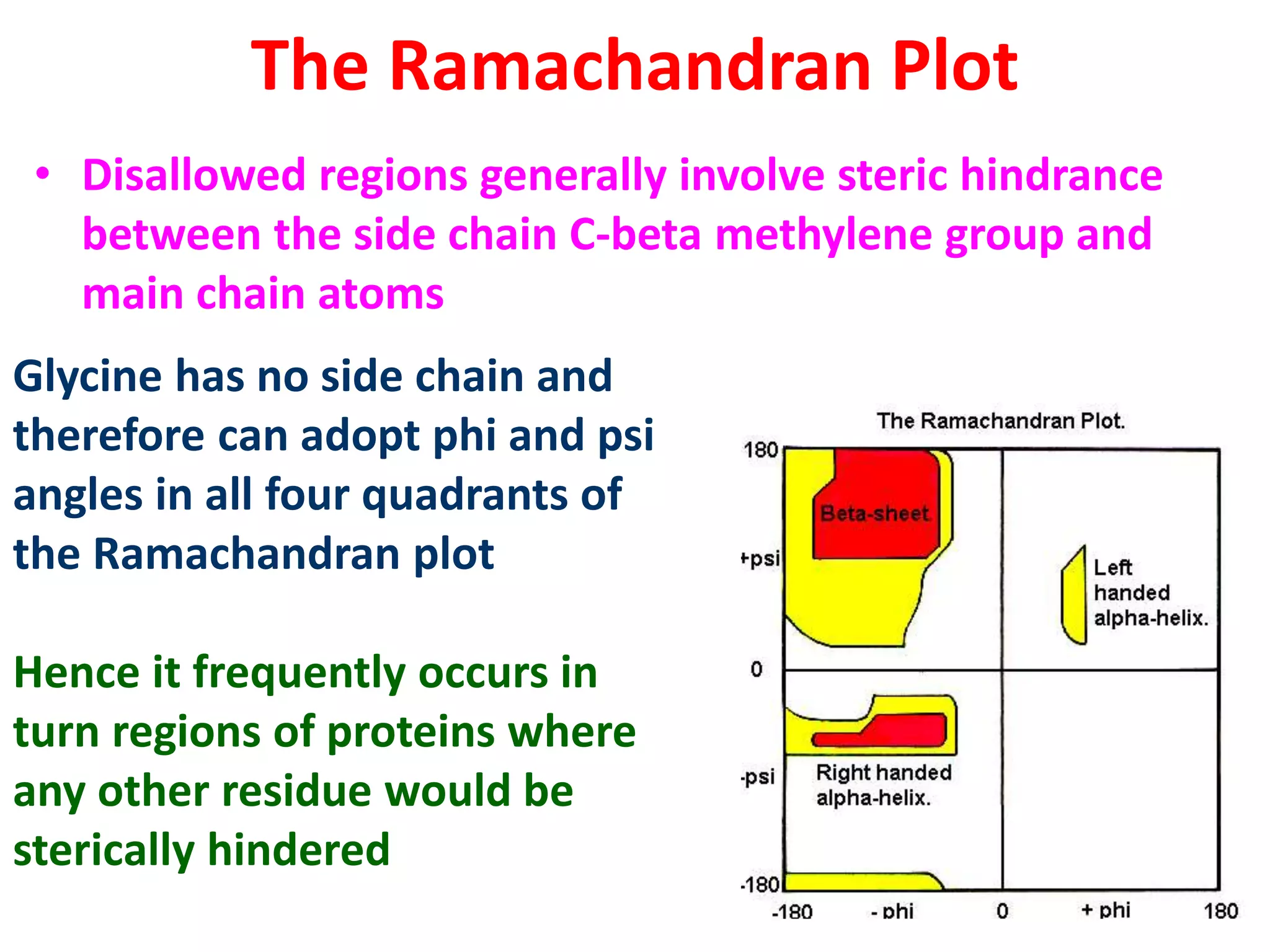
The Ramachandran plot maps the allowed regions of phi and psi dihedral angles in protein backbone structures. It revealed that phi and psi angles are restricted to specific regions to avoid steric clashes between atoms. The main allowed regions correspond to alpha-helical and beta-sheet conformations. The plot is an important tool for analyzing protein structures and validating predicted folds based on expected phi and psi angles.