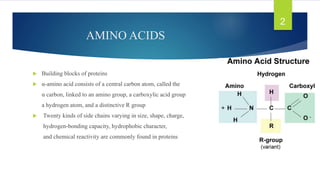
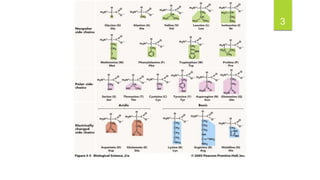
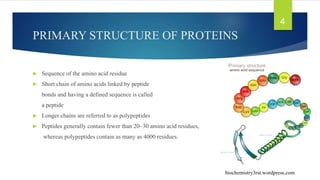
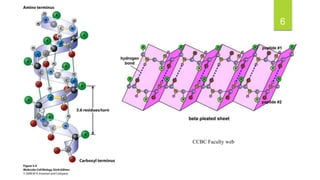
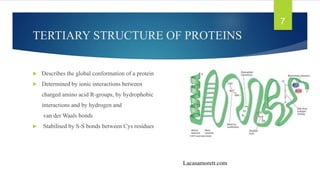
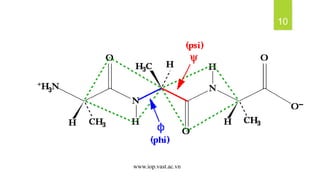
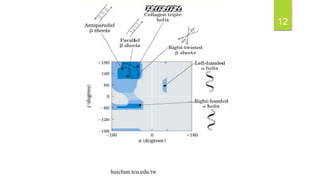
The document discusses the structure of proteins, including their primary, secondary, tertiary, and quaternary formations, indicating how amino acids and their interactions determine these structures. It also outlines the Ramachandran plot, a tool used to visualize dihedral angles of amino acid residues, highlighting allowable and disallowed angles due to steric clashes. Key points include the specific roles of amino acids like proline and glycine in the structural integrity and flexibility of proteins.