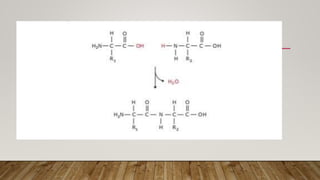
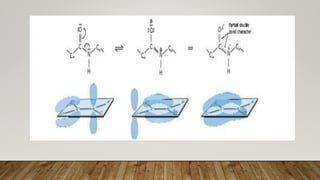
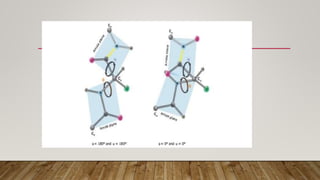
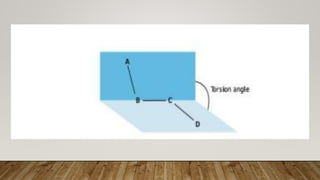
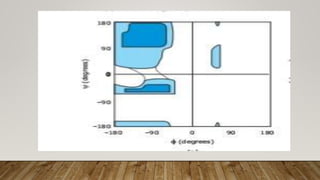
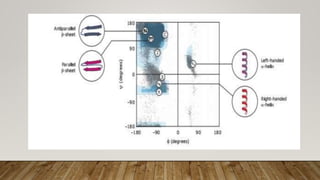
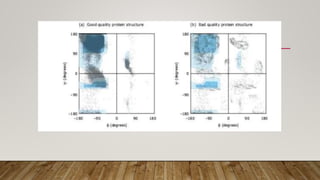
The Ramachandran plot provides a graphical representation of allowed phi and psi dihedral angles for protein backbone structures. It revealed that most phi-psi combinations are sterically prohibited except for certain allowed regions. The plot remains an important tool for validating protein structures determined experimentally or through modeling, as structures with residues falling in forbidden regions likely have low quality. It also allows prediction of secondary structure based on characteristic phi-psi angles of different structures like alpha helices and beta sheets.