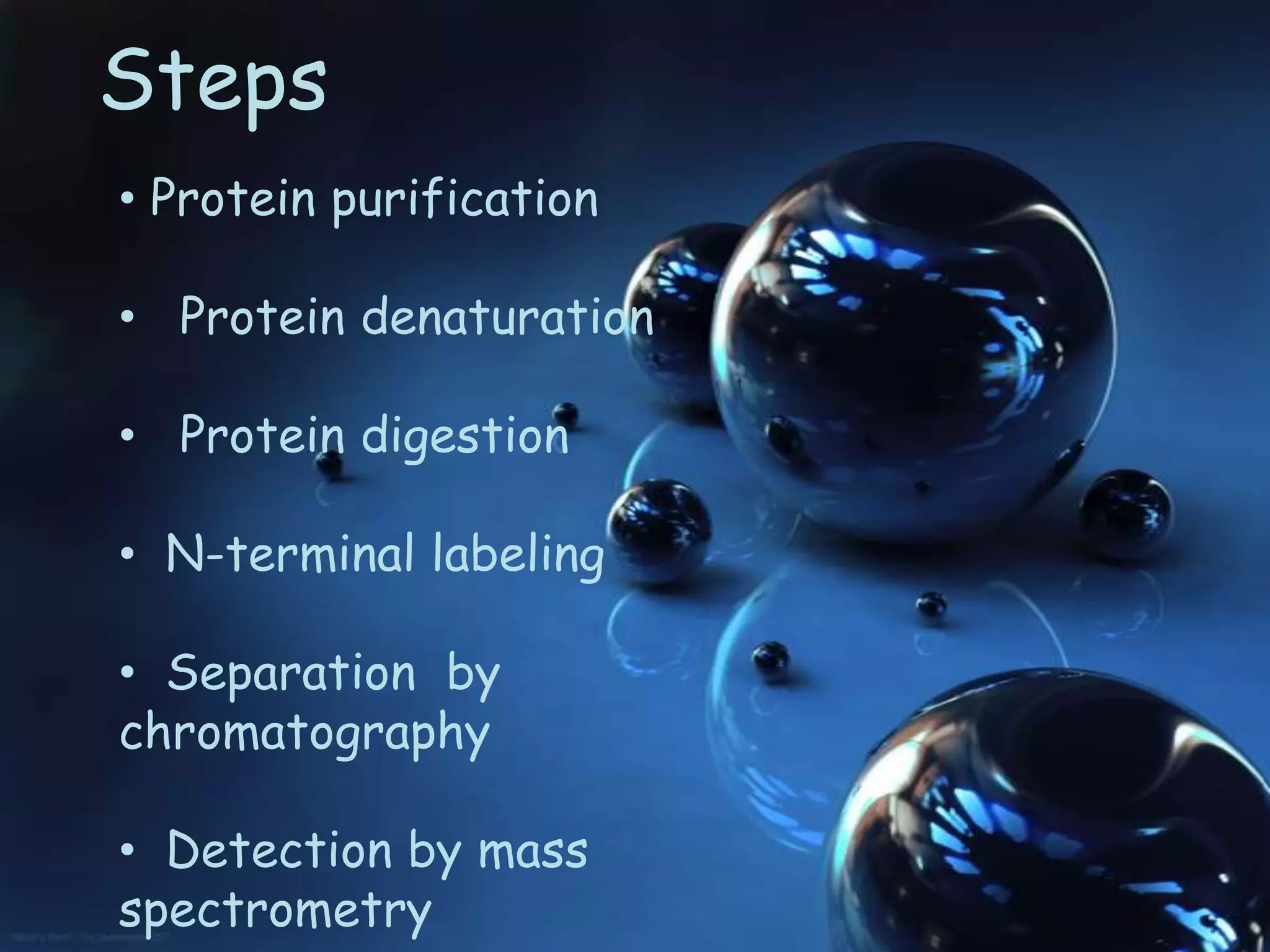
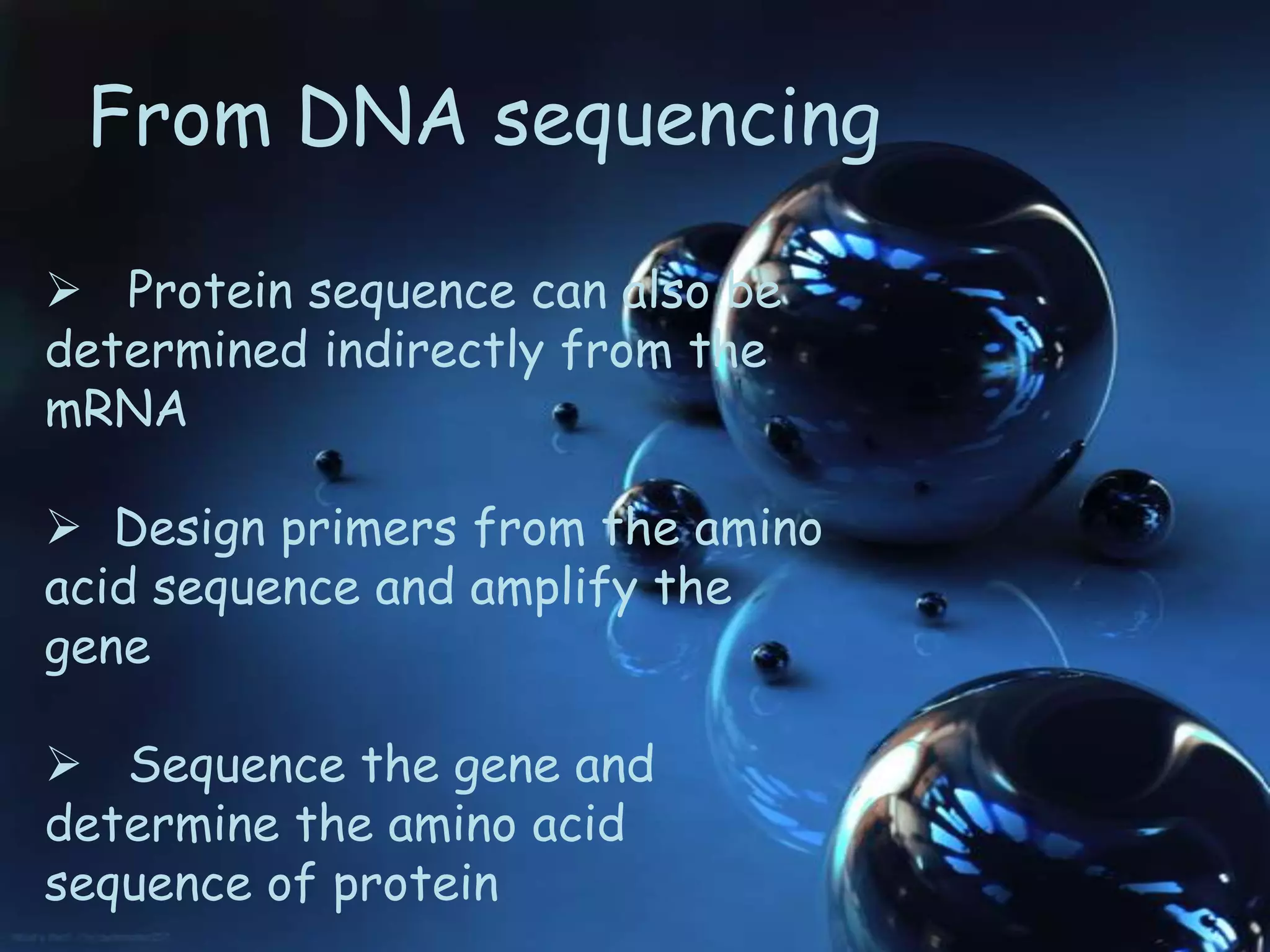
This document discusses protein sequencing techniques. There are three main methods for determining a protein's amino acid sequence: N-terminal sequencing, C-terminal sequencing, and predicting the sequence from DNA sequencing. N-terminal sequencing involves using methods like Edman degradation to sequentially remove amino acids from the N-terminus and identify them. C-terminal sequencing uses carboxypeptidases to sequentially remove amino acids from the C-terminus over time. DNA sequencing allows predicting the protein sequence by first sequencing the gene that encodes the protein. Protein sequencing is important for understanding cellular processes and developing targeted drugs.