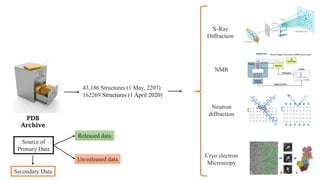
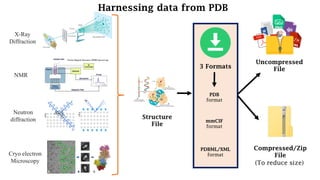
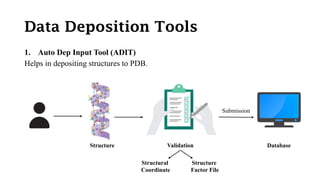
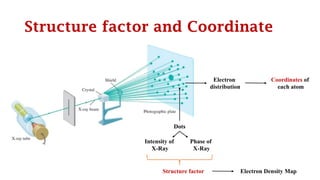
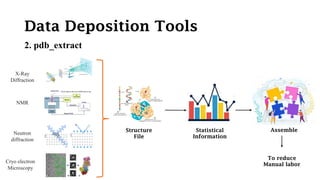
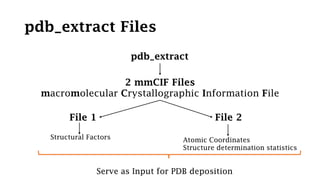
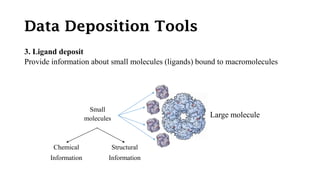
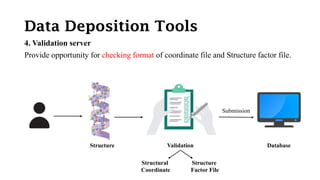
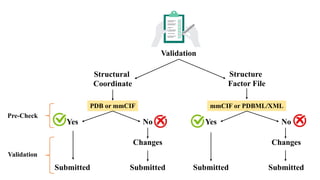
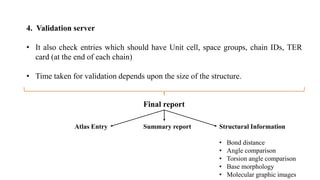
The Protein Data Bank (PDB) serves as an archive for the 3D structures of biological macromolecules, established in 1971 by the RCSB. It contains detailed experimental data, including atomic coordinates and various structural factors, with over 162,000 structures available as of April 2020. Various data deposition and validation tools assist researchers in submitting and verifying structural data, ensuring accuracy and accessibility for scientific use.