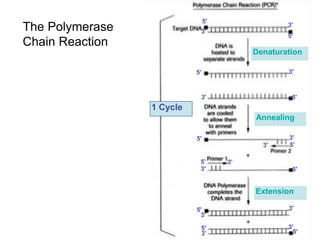
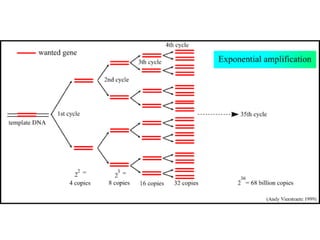
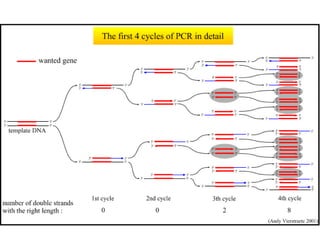
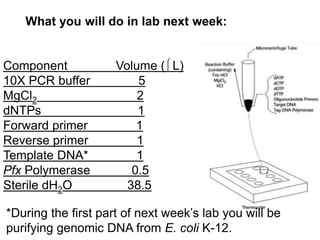
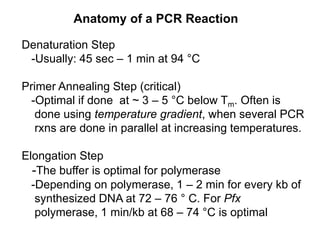
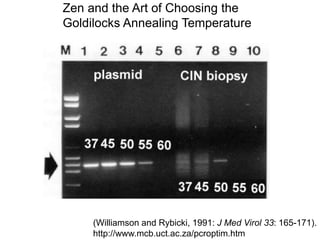
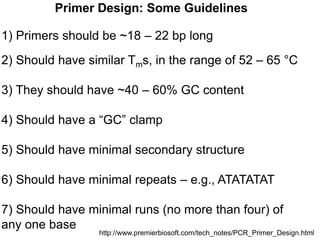
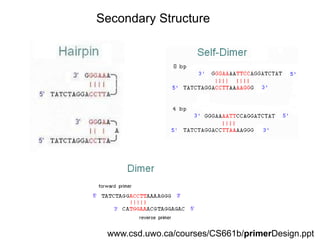
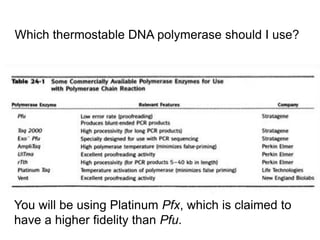
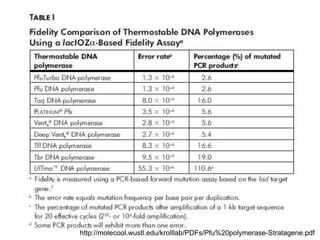
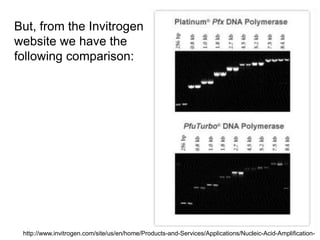
This document summarizes the invention and development of the polymerase chain reaction (PCR) technique by Kary Mullis in 1983. It provides details on how to perform PCR, including reaction components, cycling conditions, primer design guidelines, and choosing a thermostable DNA polymerase. PCR is a method to amplify a specific DNA sequence using repeated cycles of heating and cooling.