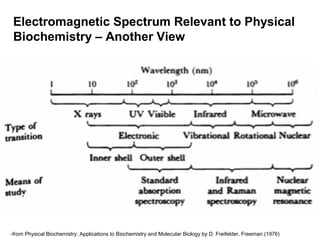
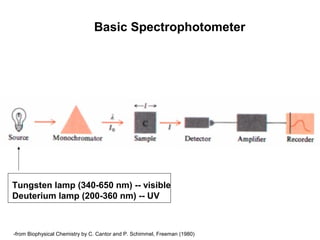
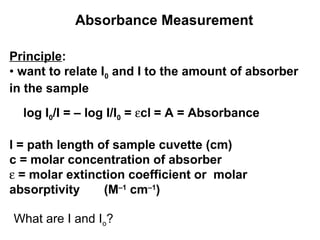
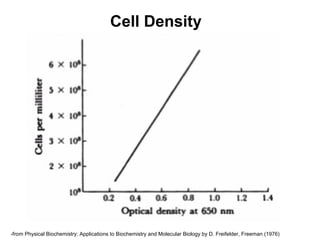
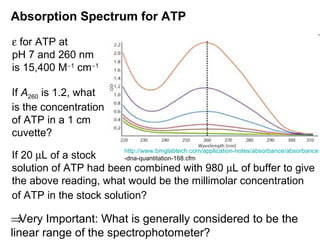
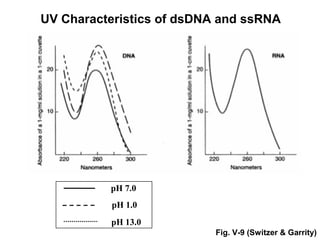
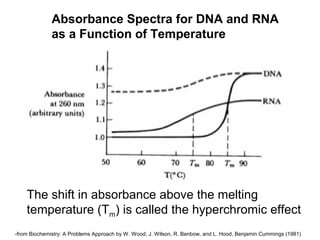
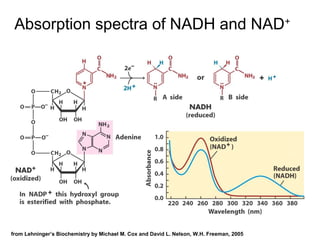
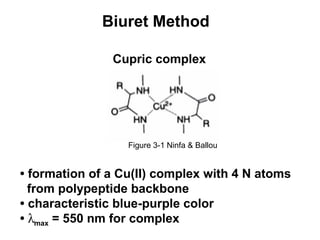
This document discusses spectrophotometry techniques for measuring light absorption by molecules. It covers the electromagnetic spectrum, Beer-Lambert law, applications of UV-vis spectroscopy like determining cell density and protein concentration, and methods for measuring absorbance of molecules like DNA, RNA, proteins, and other biological compounds. Key concepts explained include the relationship between absorbance, molar extinction coefficient, concentration, and path length in the Beer-Lambert law.


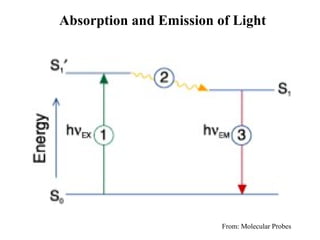
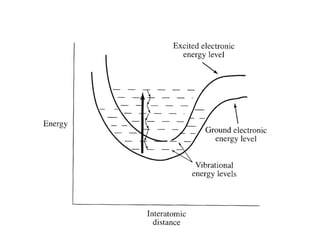
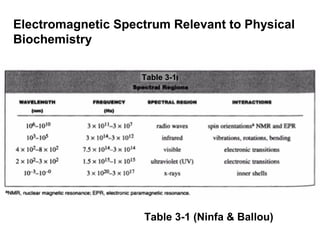




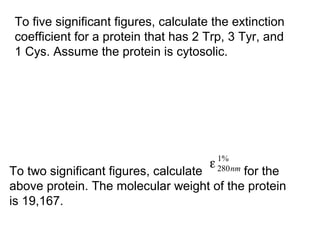
![Using A280 readings to determine the concentration
of a protein…but first we have to get a good estimate
of the molar extinction coefficient of the protein. So…
How to estimate the molar extinction coefficient (e) of
a protein:
e280nm [M-1 cm-1] = 5500×nTrp + 1490×nTyr + 125×nCystines
Need sequence information to use this method, and
if you know if the protein has disulfides, the estimate
improves. (We will talk more about this in class.)
This empirical equation is taken from: Pace et al.
(1995) Protein Science 4, 2411](https://image.slidesharecdn.com/spectrophotometrylecture-140903124737-phpapp02/85/Spectrophotometry-Lecture-27-320.jpg)