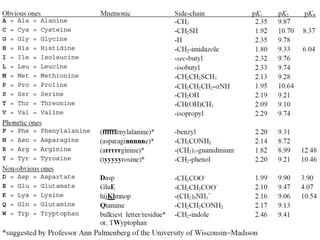
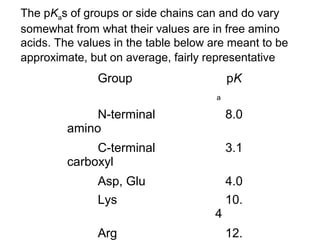
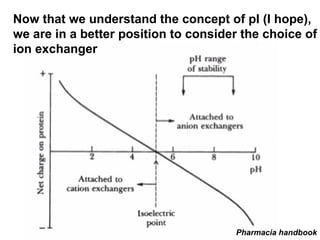
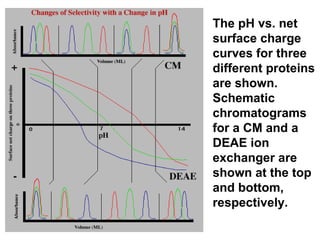
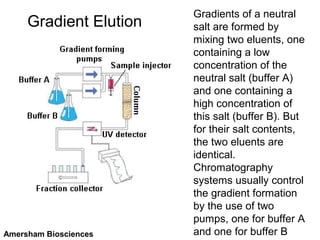
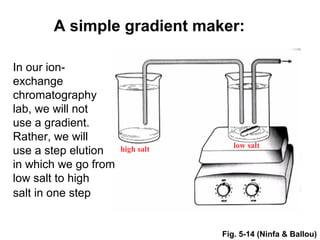
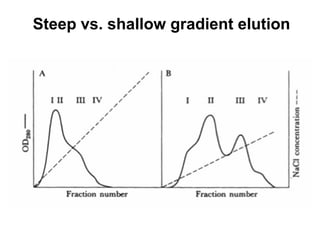
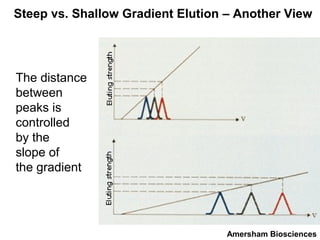
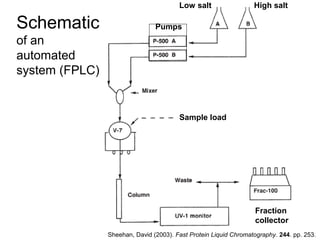
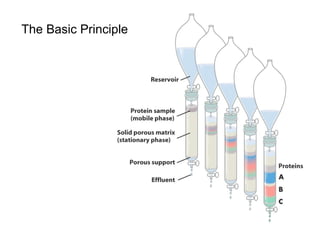
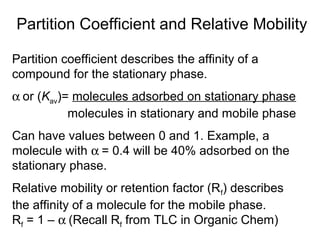
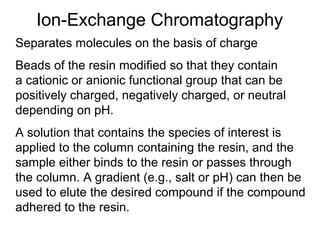
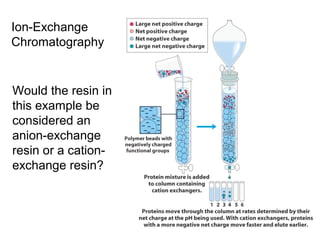
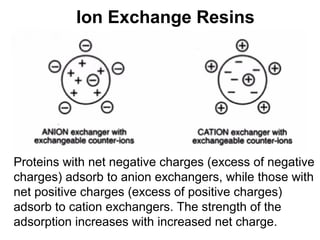
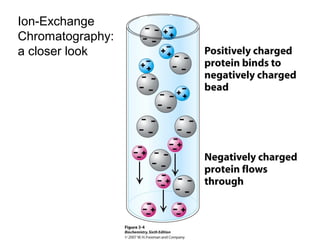
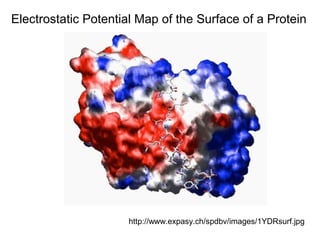
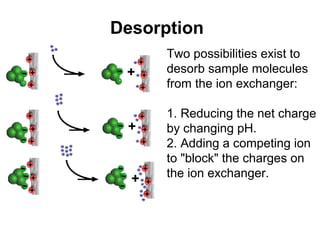
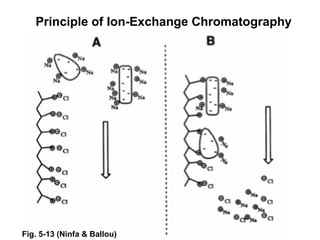
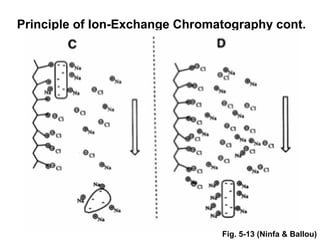
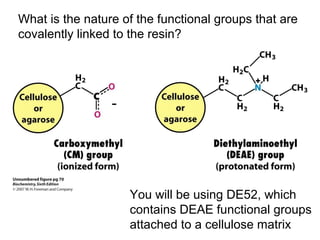
Ion-exchange chromatography separates molecules on the basis of charge. The stationary phase contains resin beads with cationic or anionic functional groups that can bind positively or negatively charged molecules. Proteins either bind to the resin based on their net charge or pass through. Elution is achieved by changing pH or adding salts to compete for binding sites on the resin. Modern systems automate ion-exchange chromatography using gradients controlled by multiple pumps and fraction collectors.



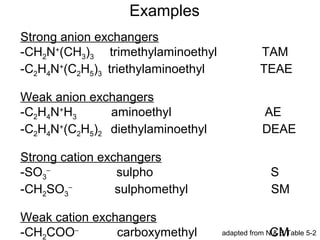
![How Do We Know If “Our” Protein Is Going
to Bind the Ion-Exchange Resin That We Are
Using? – pH, pKa, pI & Buffers Revisited:
pH = -log[H+] (not strictly true but a useful, working
definition)
pH = pKa + log([basic form]/[acidic form]) [HH eq]
Isoelectric point (pI) is the pH at which a molecule
has a net charge of zero.
Buffers useful ±1 (or ±0.5) units above and below
their pKa](https://image.slidesharecdn.com/ionexchangechromatographylecture-140903125126-phpapp02/85/Ion-Exchange-Chromatography-Lecture-16-320.jpg)