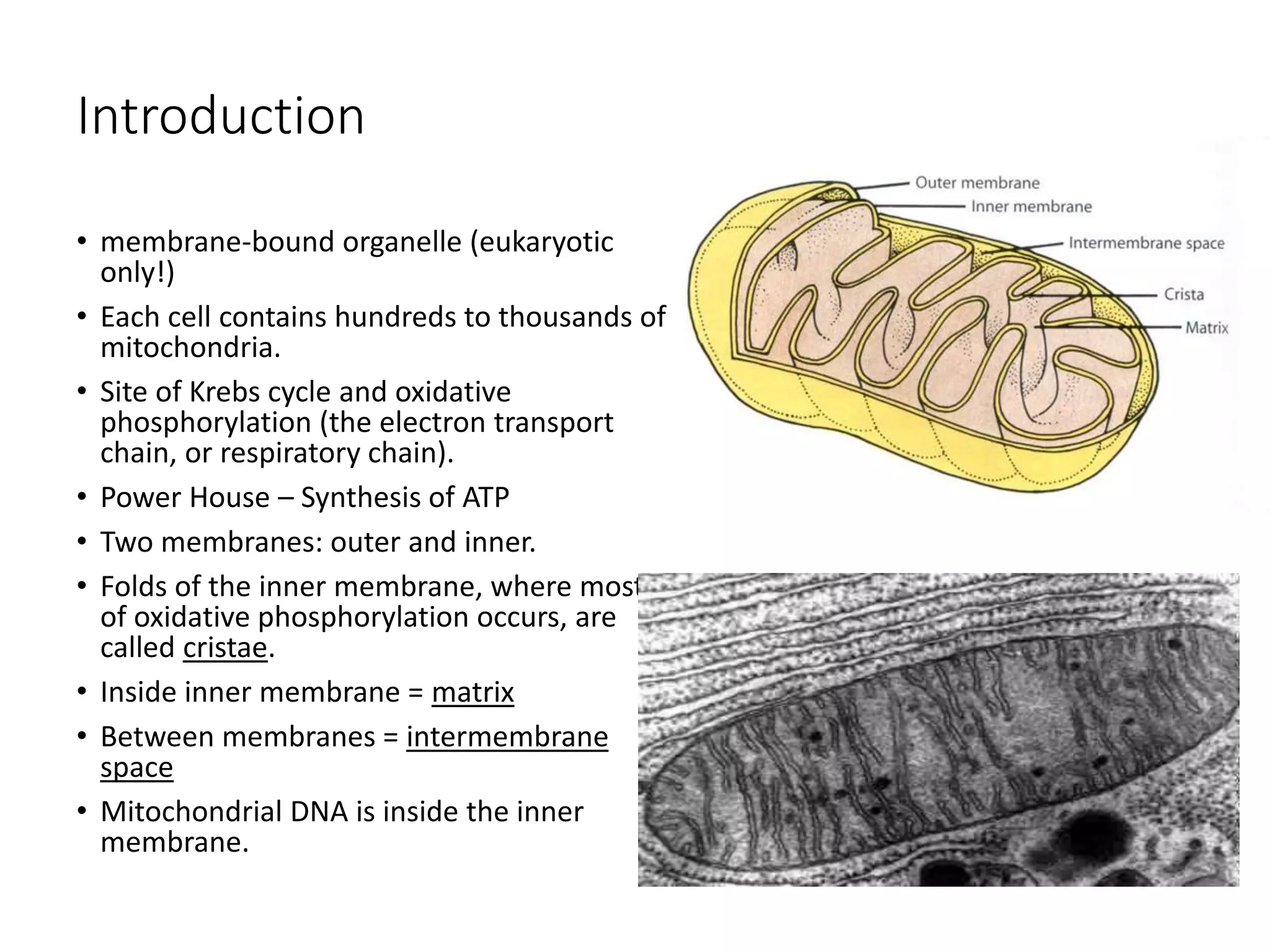
The document provides an overview of mitochondrial genomes, detailing their structure, function, and evolutionary relevance based on the endosymbiont hypothesis. It highlights mitochondrial processes such as the Krebs cycle, electron transport, and unique features like the presence of ribosomal RNA and genes for respiratory functions. Additionally, it discusses the complexity of plant mitochondrial DNA and its interactions with chloroplast DNA.