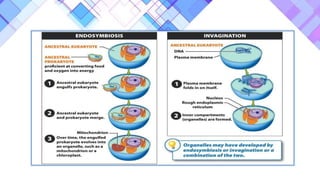
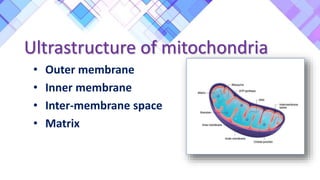
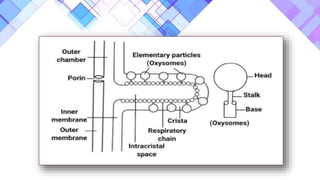
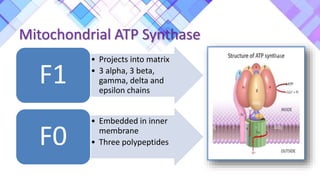
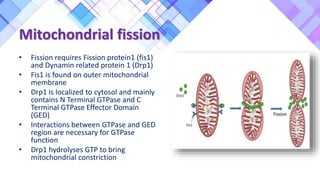
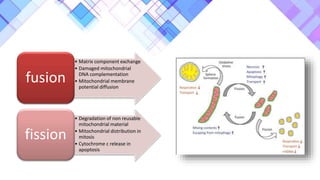
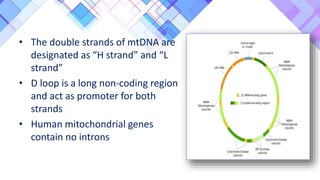
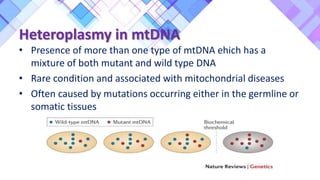
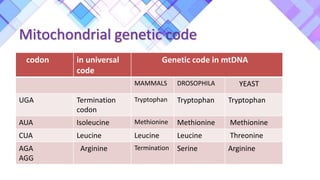
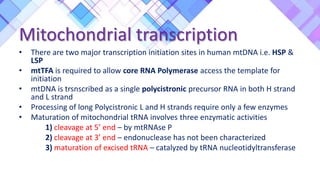
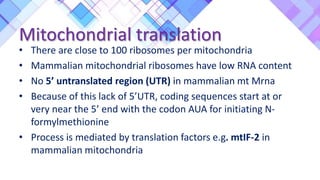
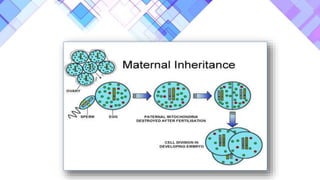
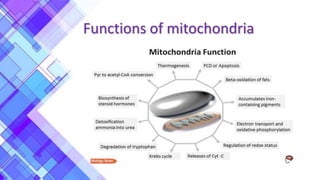
The document provides a comprehensive overview of mitochondria, including their structure, history, genetic characteristics, and functions within eukaryotic cells. It discusses mitochondrial dynamics, replication, transcription, translation, and inheritance, as well as implications for aging and diseases related to mitochondrial dysfunction. Key points include the endosymbiotic origin theory, the significance of mitochondrial DNA, and the role of mitochondria in processes such as apoptosis and cellular metabolism.