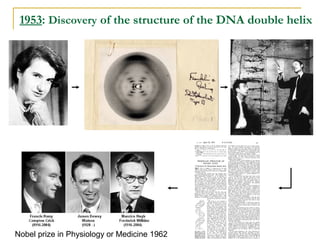
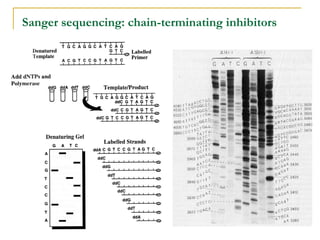
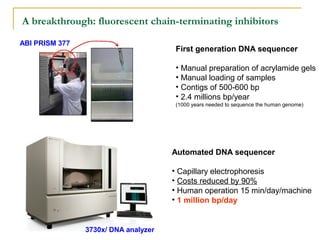
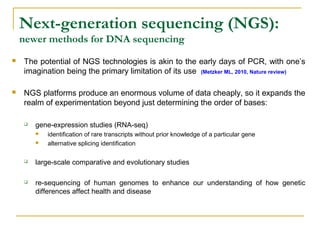
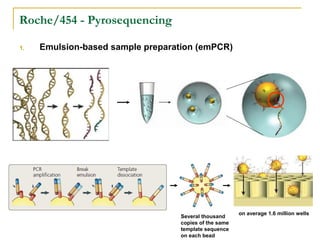
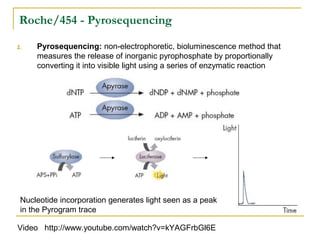
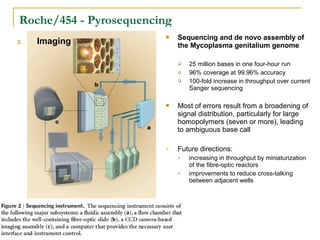
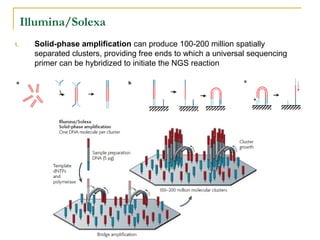
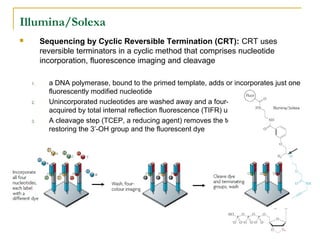
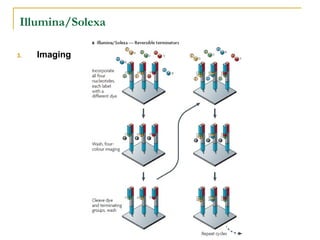
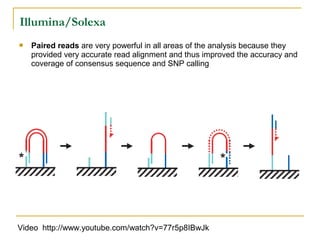
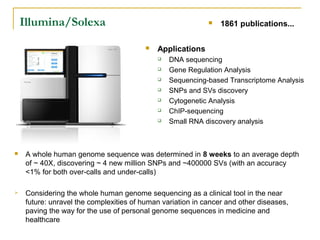
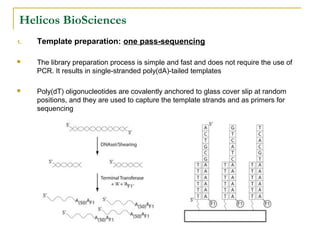
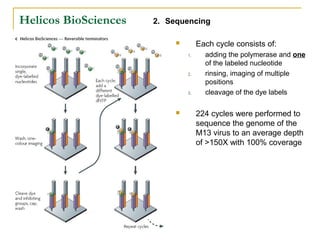
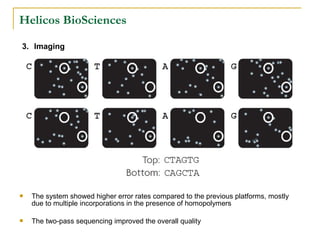
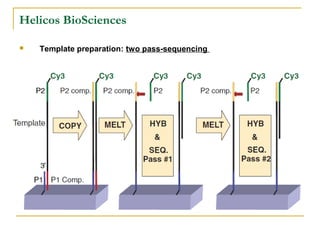
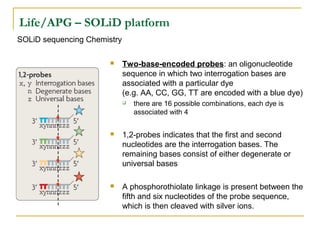
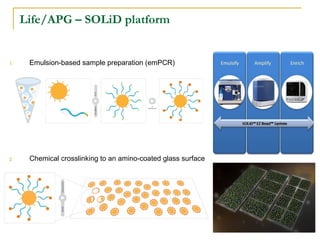
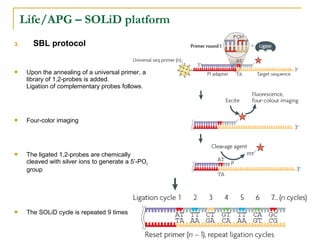
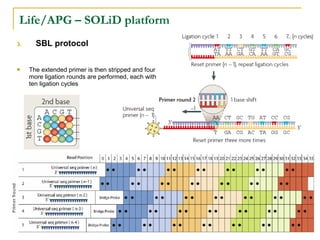
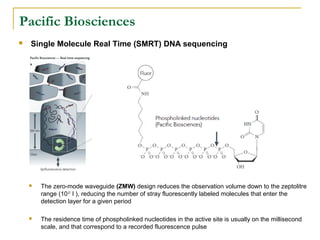
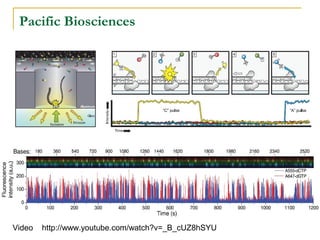
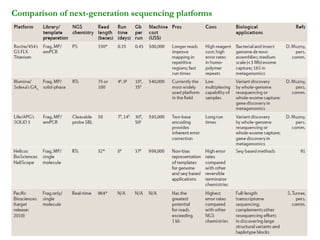
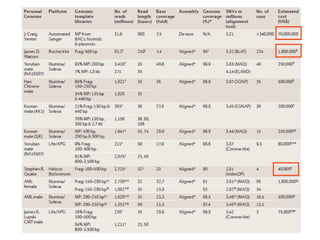
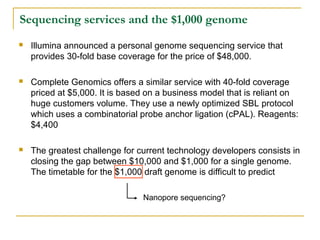
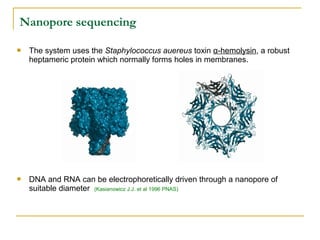
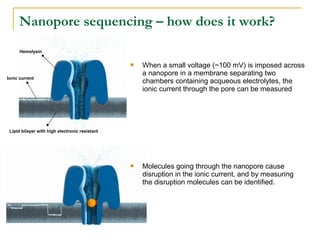
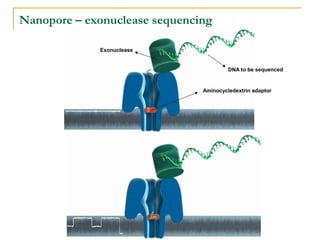
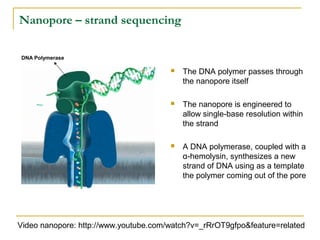
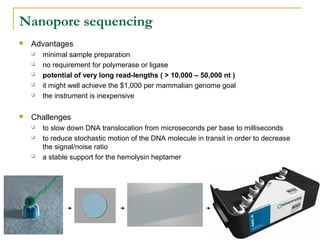
The document provides a history of DNA sequencing technologies. It begins with the discovery of DNA's structure in 1953 and the development of recombinant DNA technology in the 1970s. First generation Sanger sequencing produced short reads over 1,000 years to sequence the human genome. Next generation sequencing (NGS) platforms since 2005 have dramatically reduced costs while increasing throughput. NGS methods like Roche/454 pyrosequencing, Illumina/Solexa sequencing by synthesis, SOLiD ligation sequencing, and single-molecule real-time sequencing by Pacific Biosciences now enable large-scale genome and transcriptome analysis.