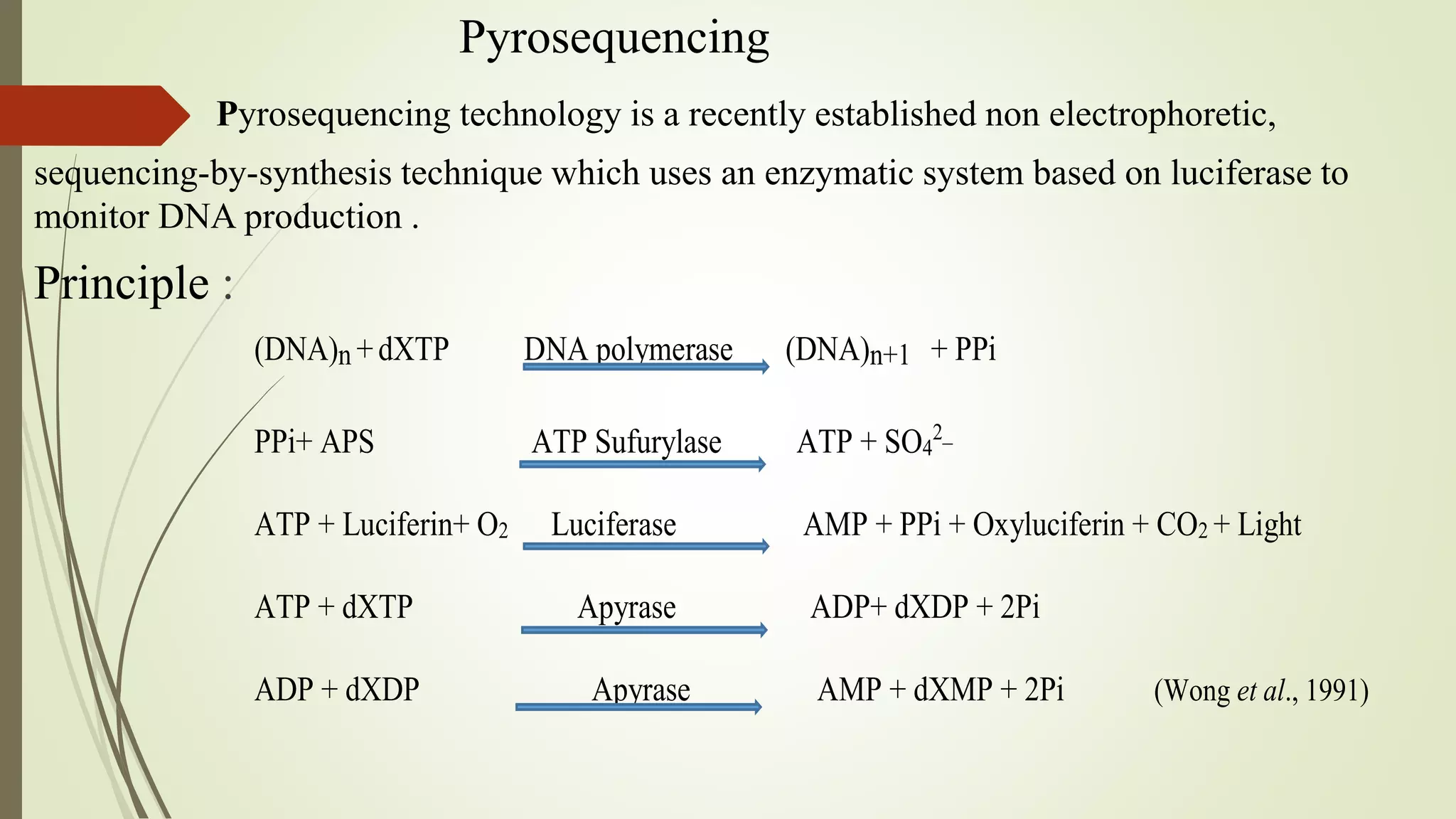
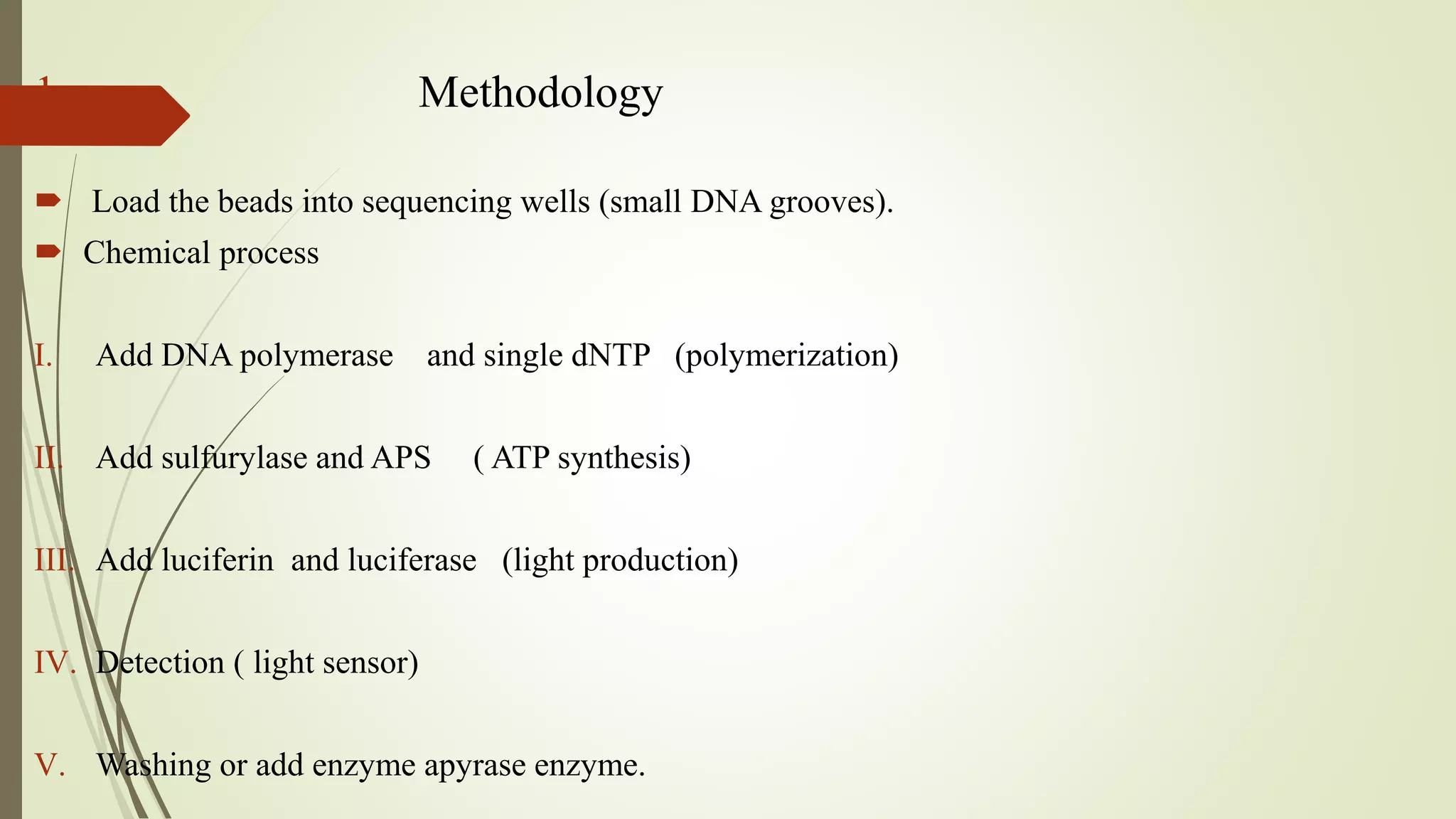
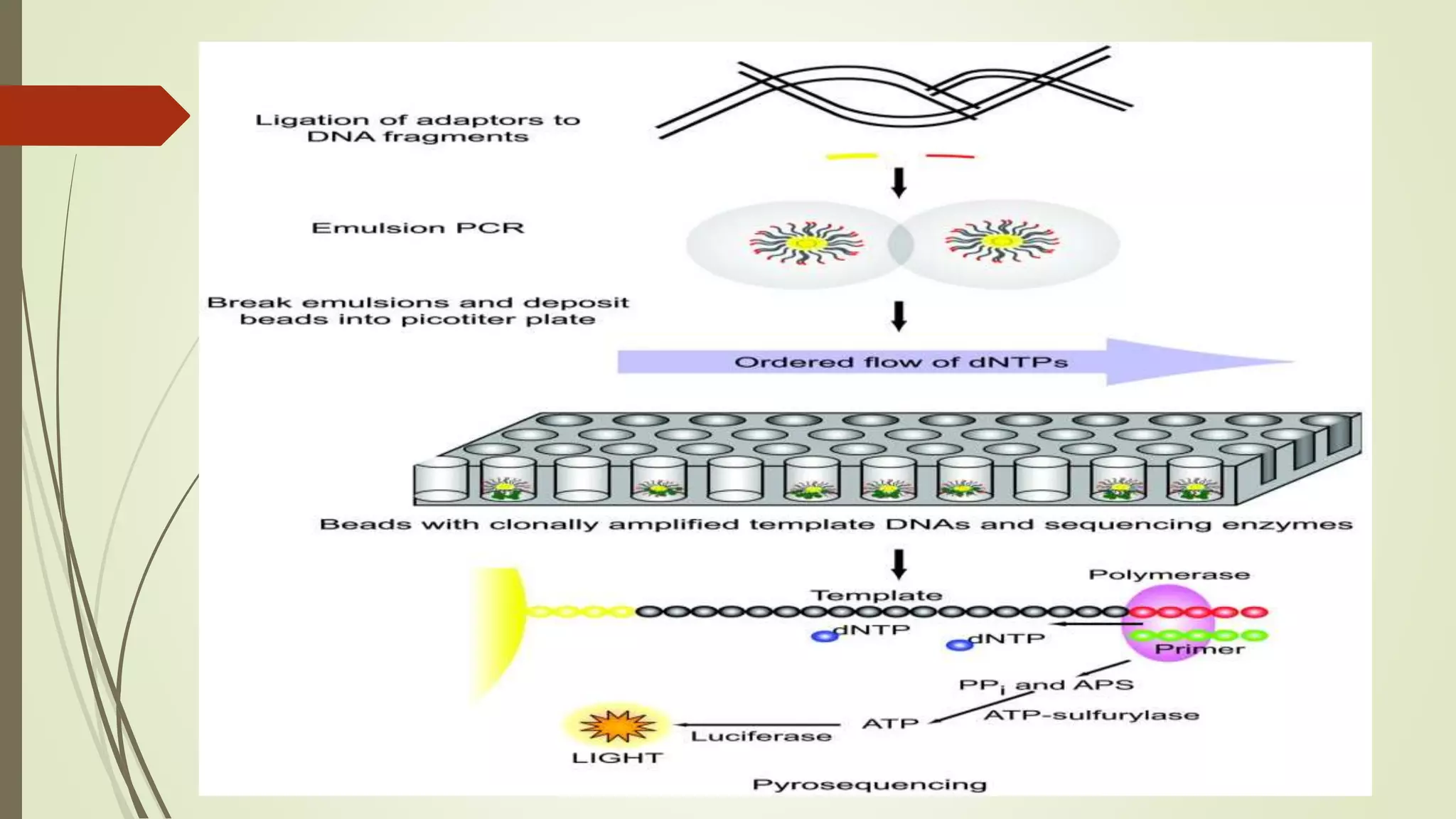
Pyrosequencing is a sequencing by synthesis technique that uses a luciferase enzyme system to monitor DNA synthesis. It works by adding DNA polymerase and a single nucleotide to the DNA fragments, generating pyrophosphate that is converted to light. The light is detected and identifies the nucleotide incorporated. Pyrosequencing has applications in cDNA analysis, mutation detection, re-sequencing of disease genes, and identifying single nucleotide polymorphisms and typing bacteria and viruses.