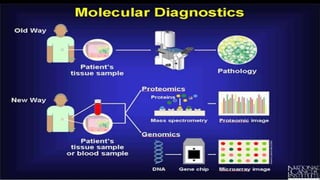
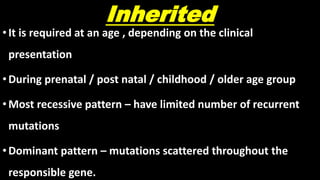
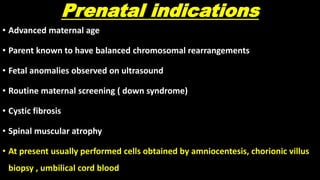
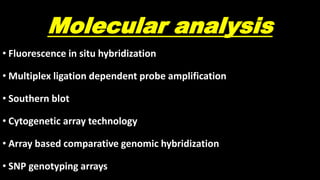
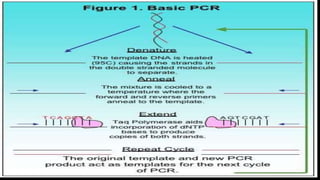
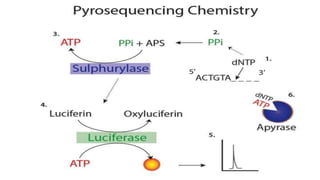
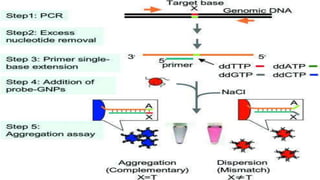
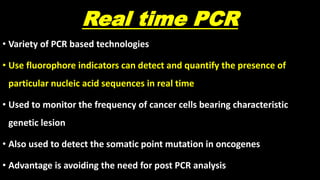
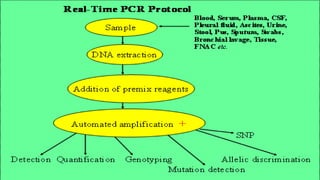
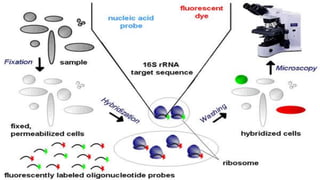
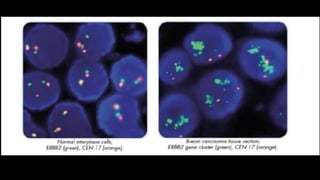
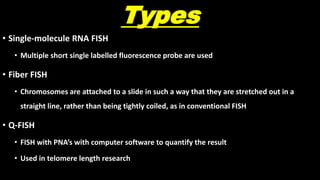
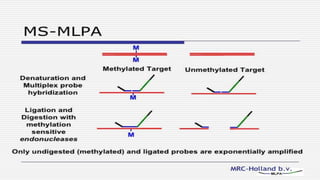
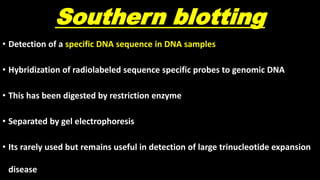
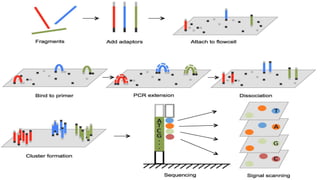
This document discusses molecular genetic diagnosis techniques. It begins by describing how techniques like karyotyping, Southern blotting, and Sanger sequencing revolutionized genetic diagnosis in the late 20th century. It then covers the various indications for genetic testing, including inherited conditions, prenatal testing, and acquired conditions like cancer. The rest of the document details specific molecular analysis techniques used, such as PCR, Sanger sequencing, pyrosequencing, restriction fragment length analysis, FISH, MLPA, Southern blotting, and next generation sequencing. It provides examples of the medical applications of these various techniques.