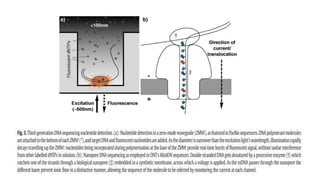
The document discusses the history and evolution of DNA sequencing technologies from the first generation, characterized by Sanger's chain termination method, to second generation techniques like pyrosequencing, and finally third generation methods including single molecule and nanopore sequencing. Key milestones include the development of various sequencing methods and their impacts on biological research, highlighted by advances such as capillary electrophoresis and the Illumina platform. The paper concludes by noting the profound implications of DNA sequencing on understanding and differentiating living organisms.