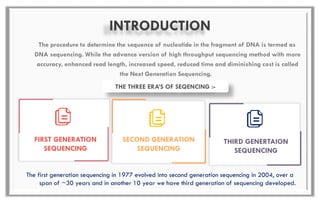
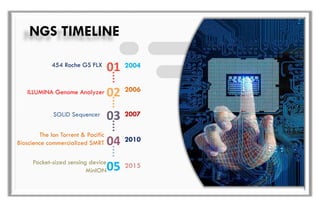
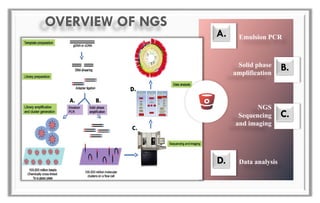
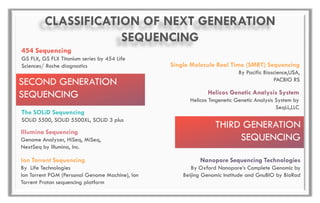
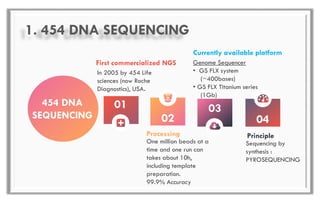
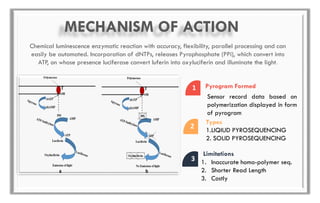
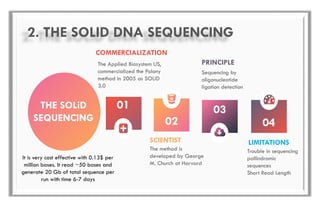
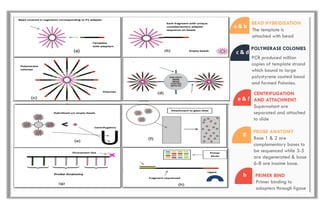
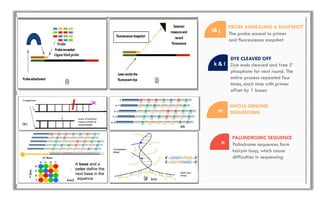
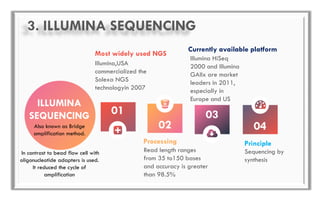
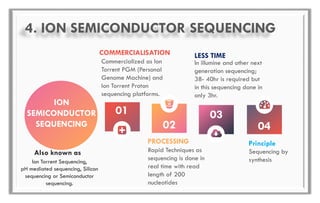
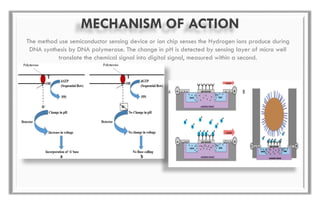
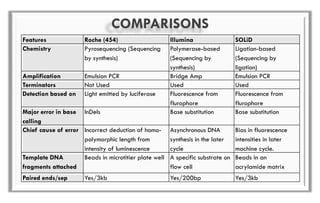
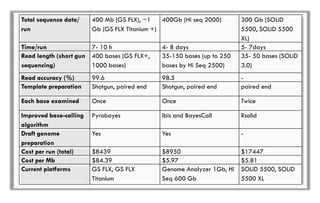
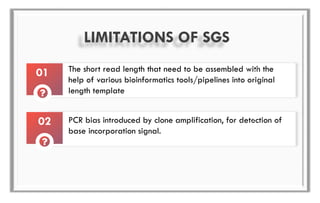
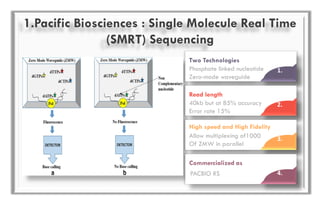
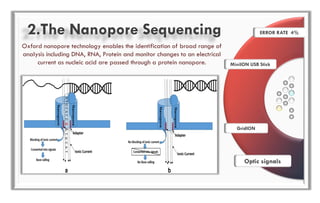
The document discusses the evolution of DNA sequencing technologies, detailing the advancements from first generation to third generation sequencing, with a focus on next generation sequencing (NGS). It highlights the advantages of NGS, such as increased speed, accuracy, reduced costs, and the ability to sequence whole genomes. Additionally, it emphasizes the implications of these advancements for fields like genetics, agriculture, and medicine.