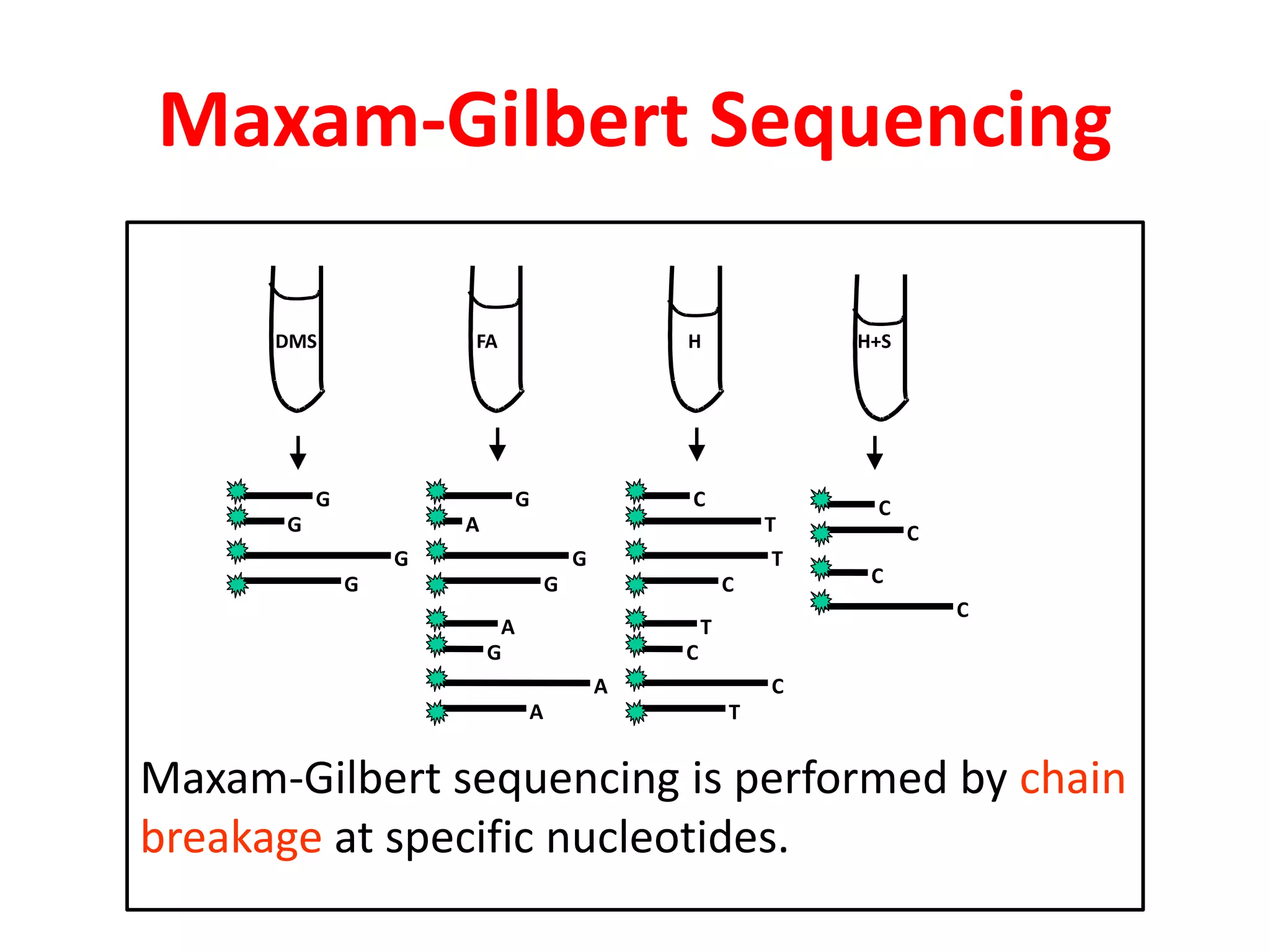
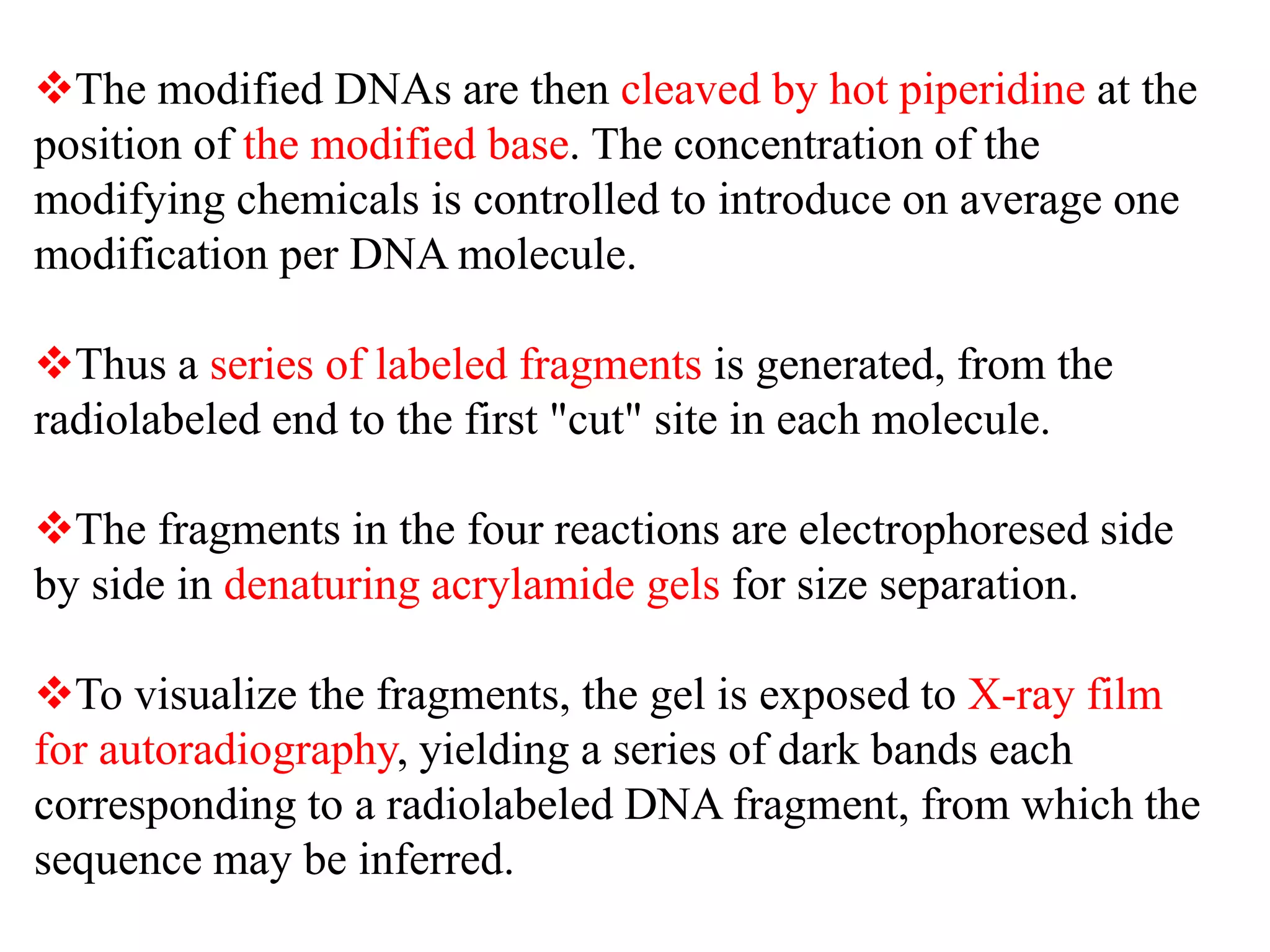
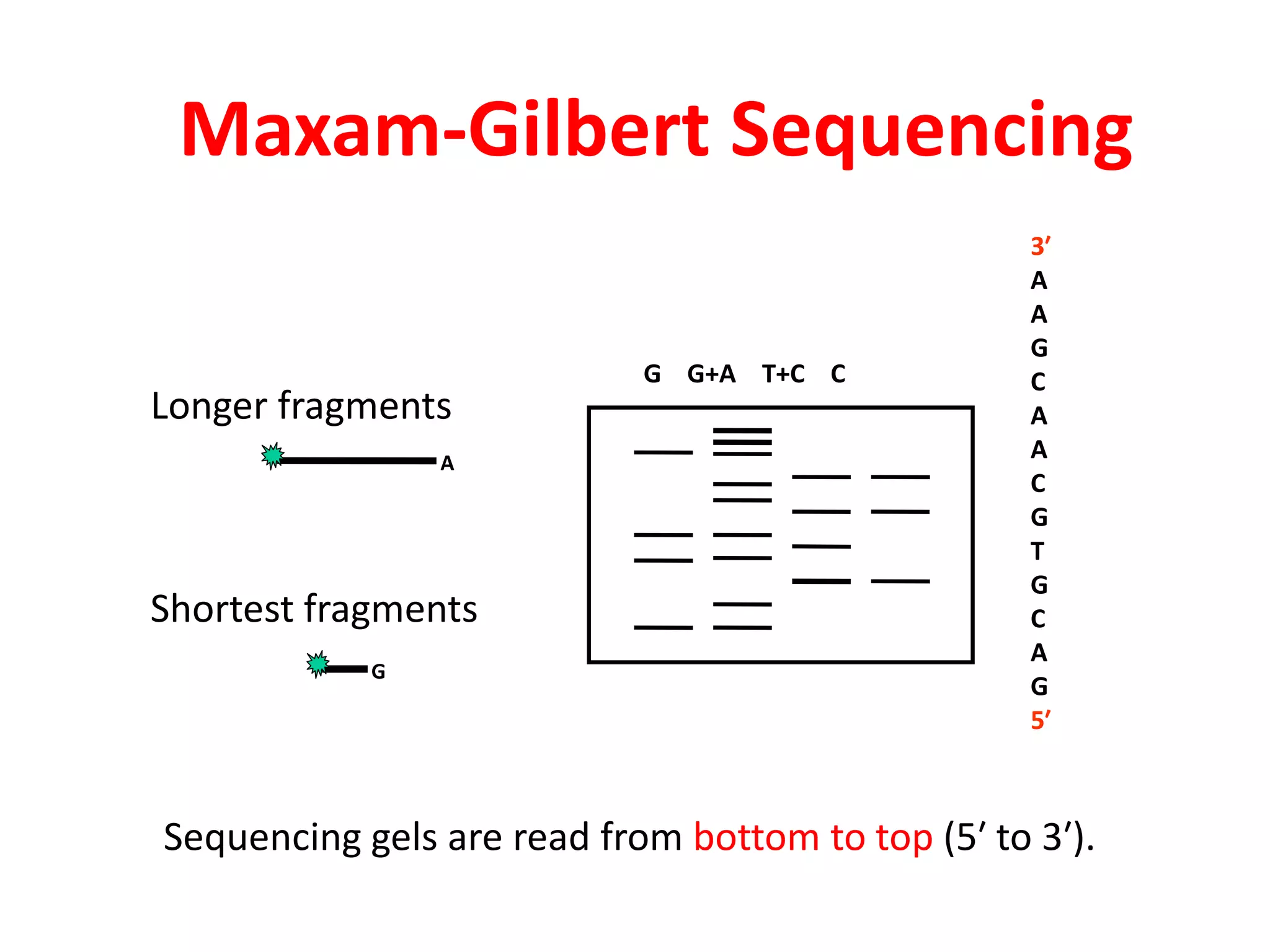
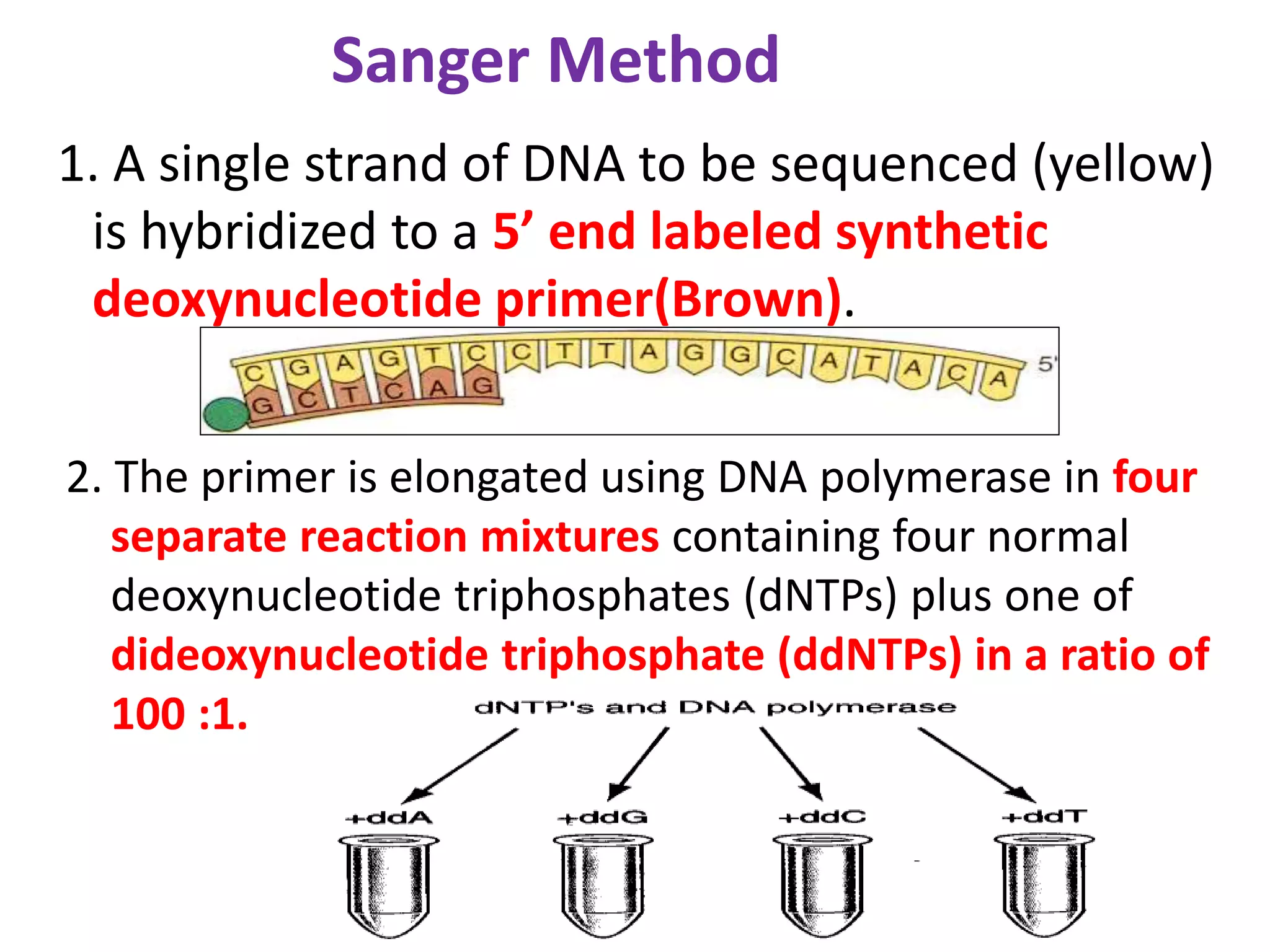
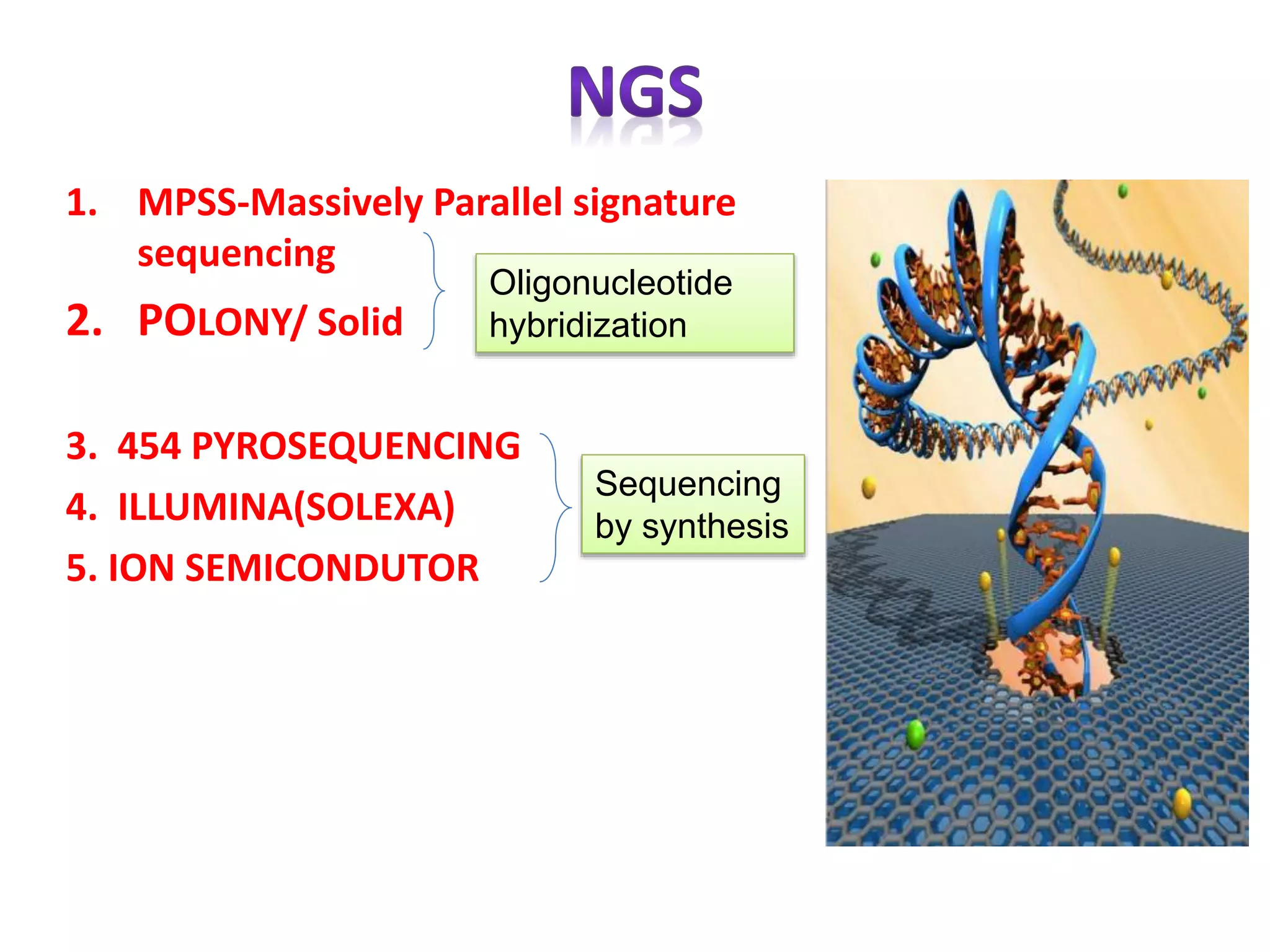
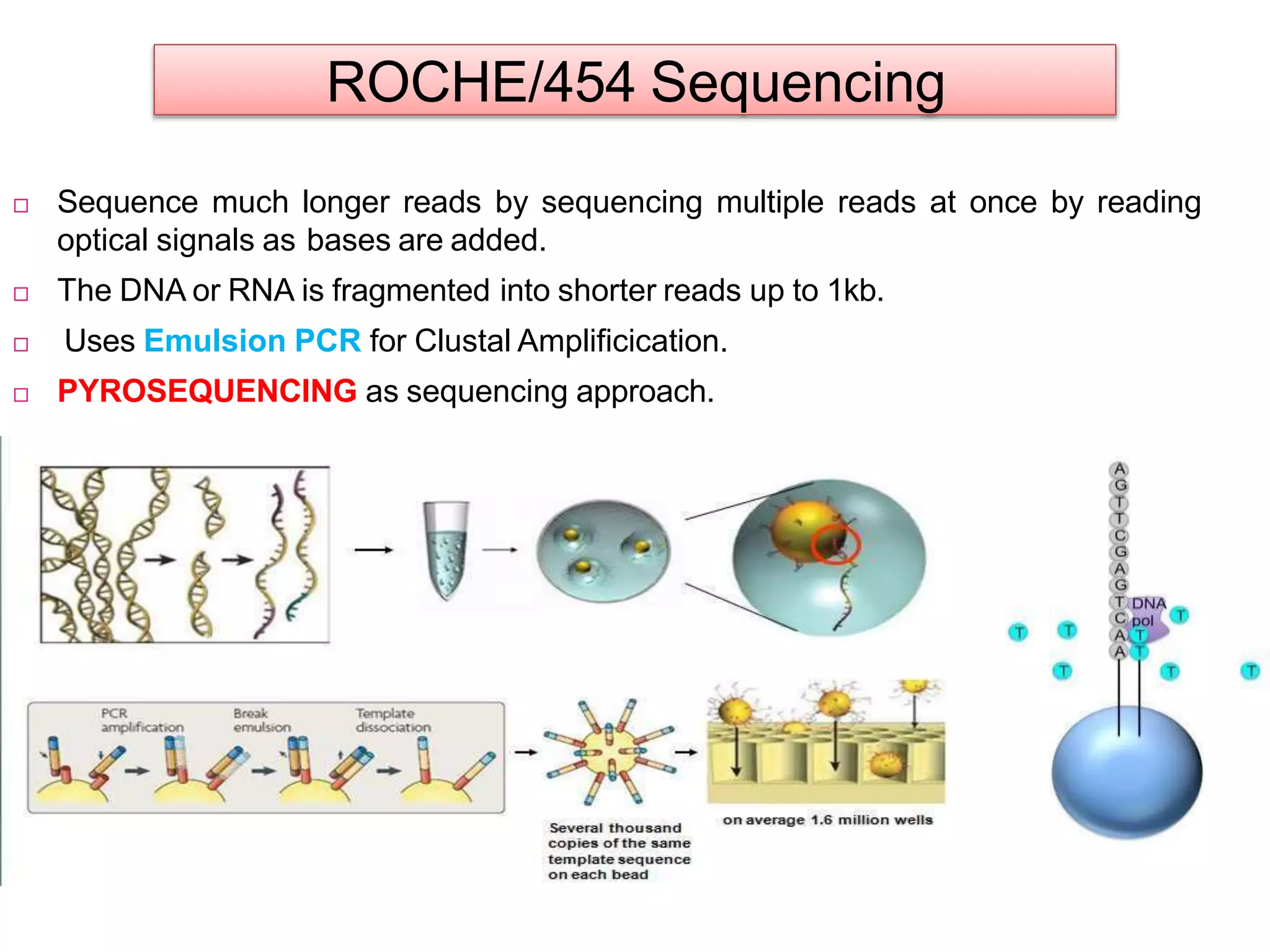
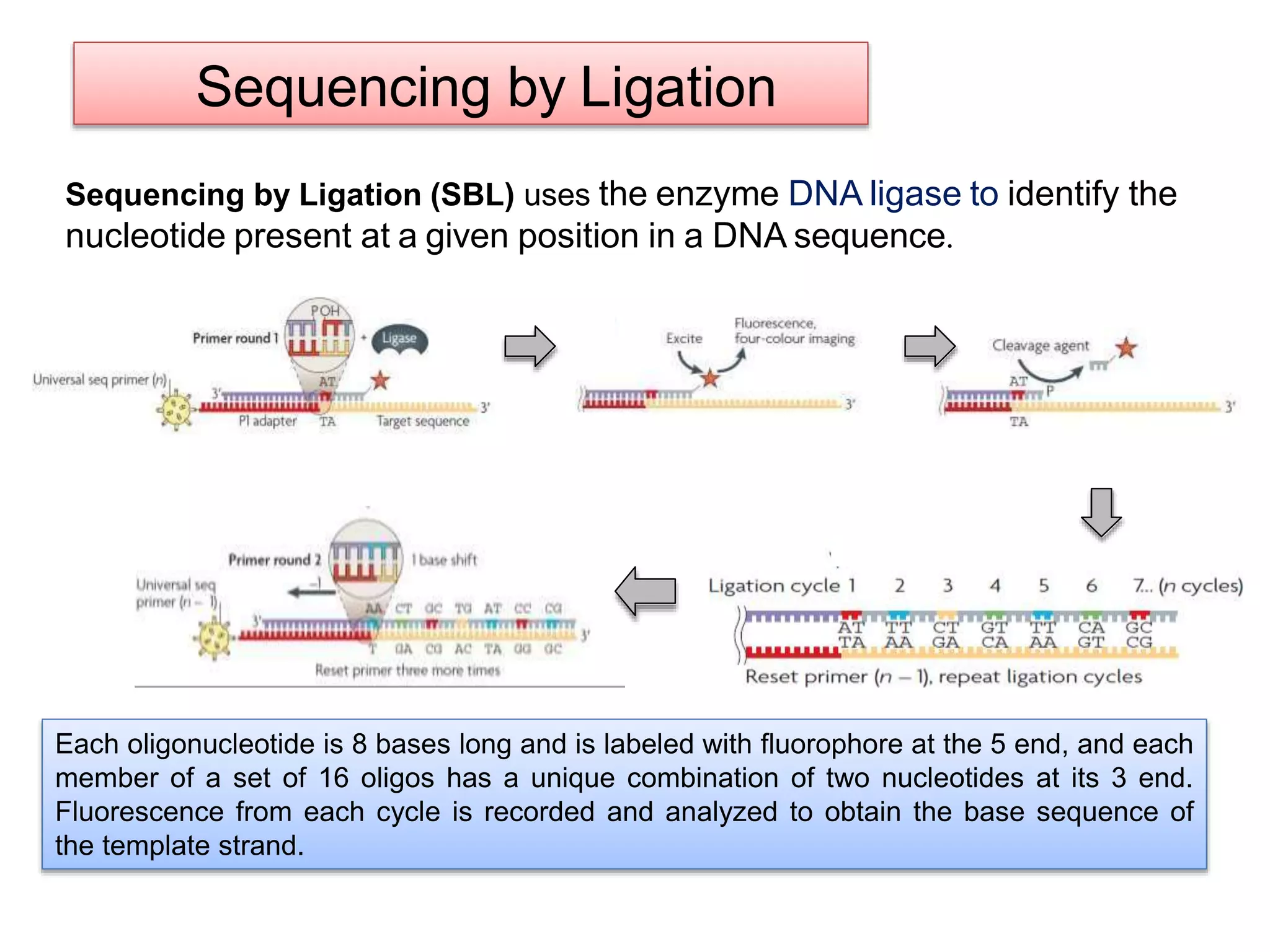
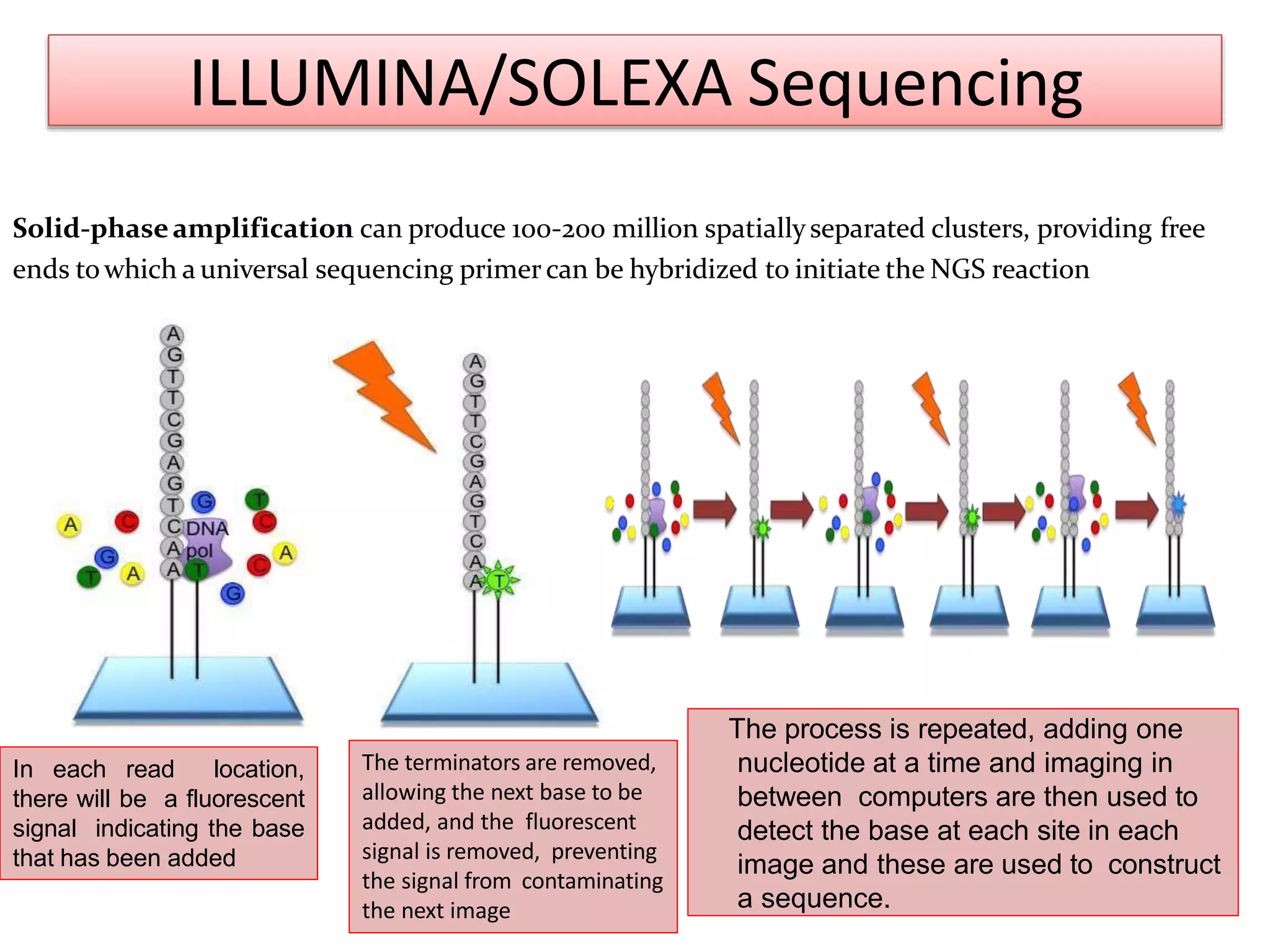
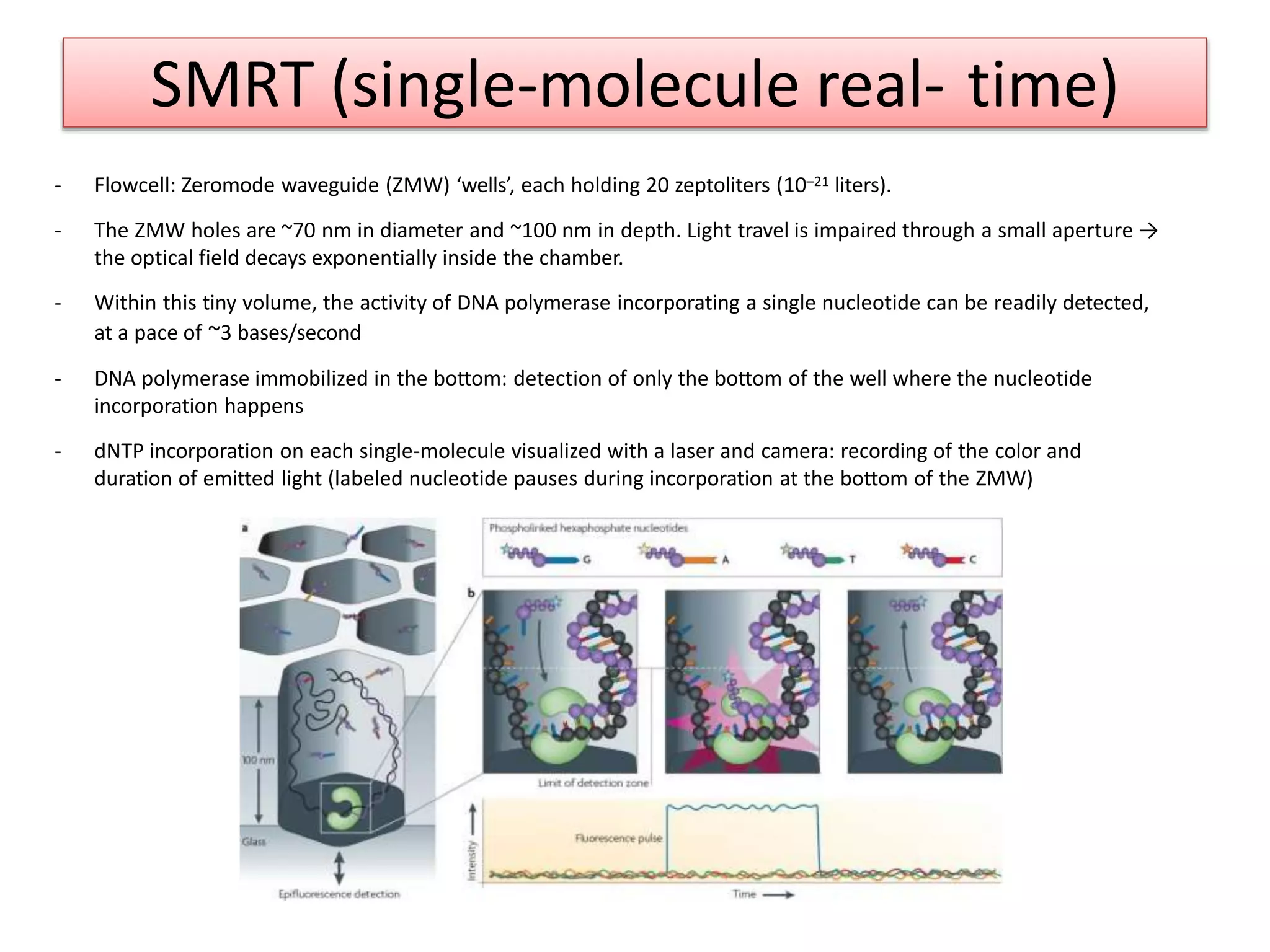
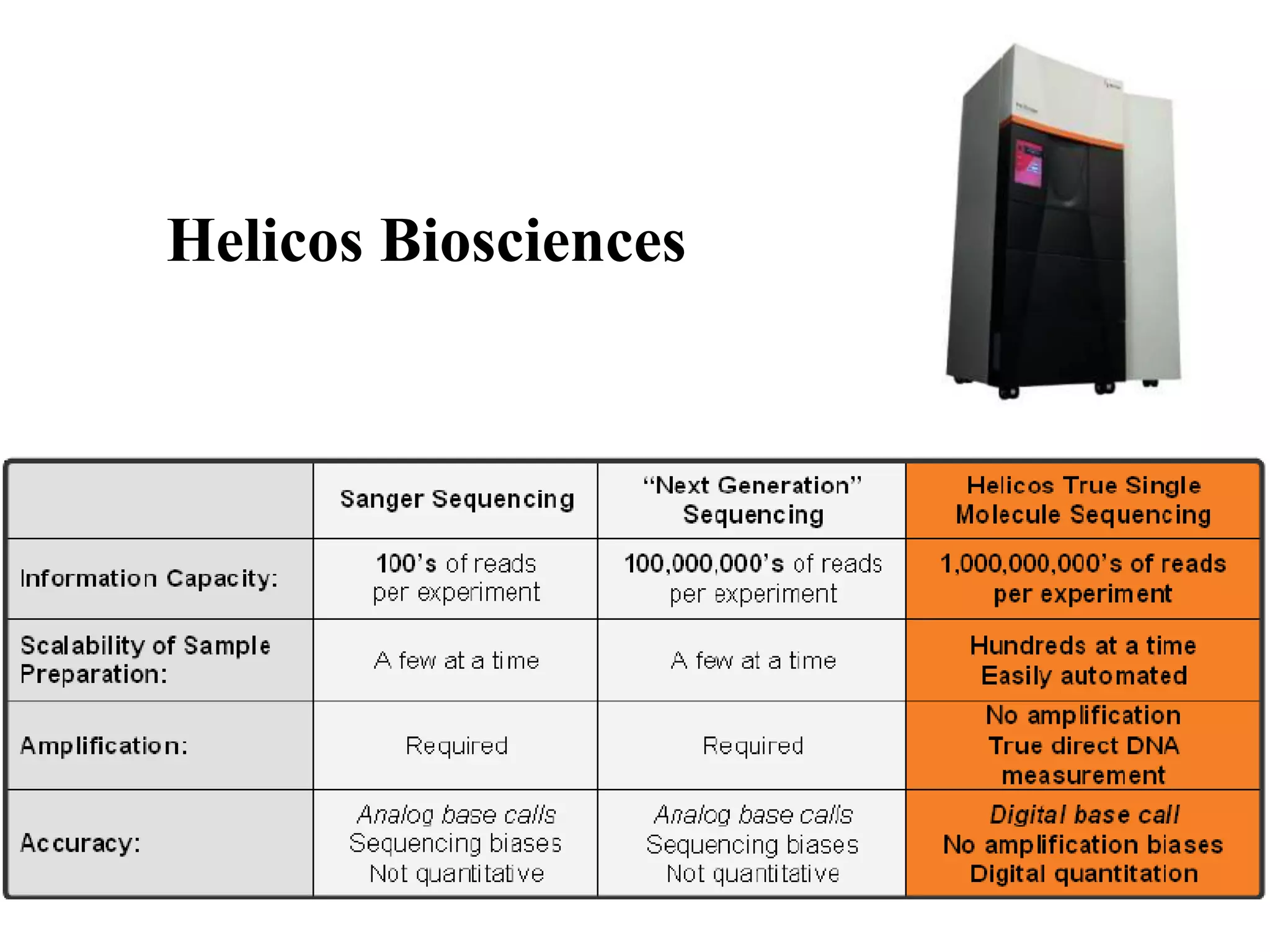
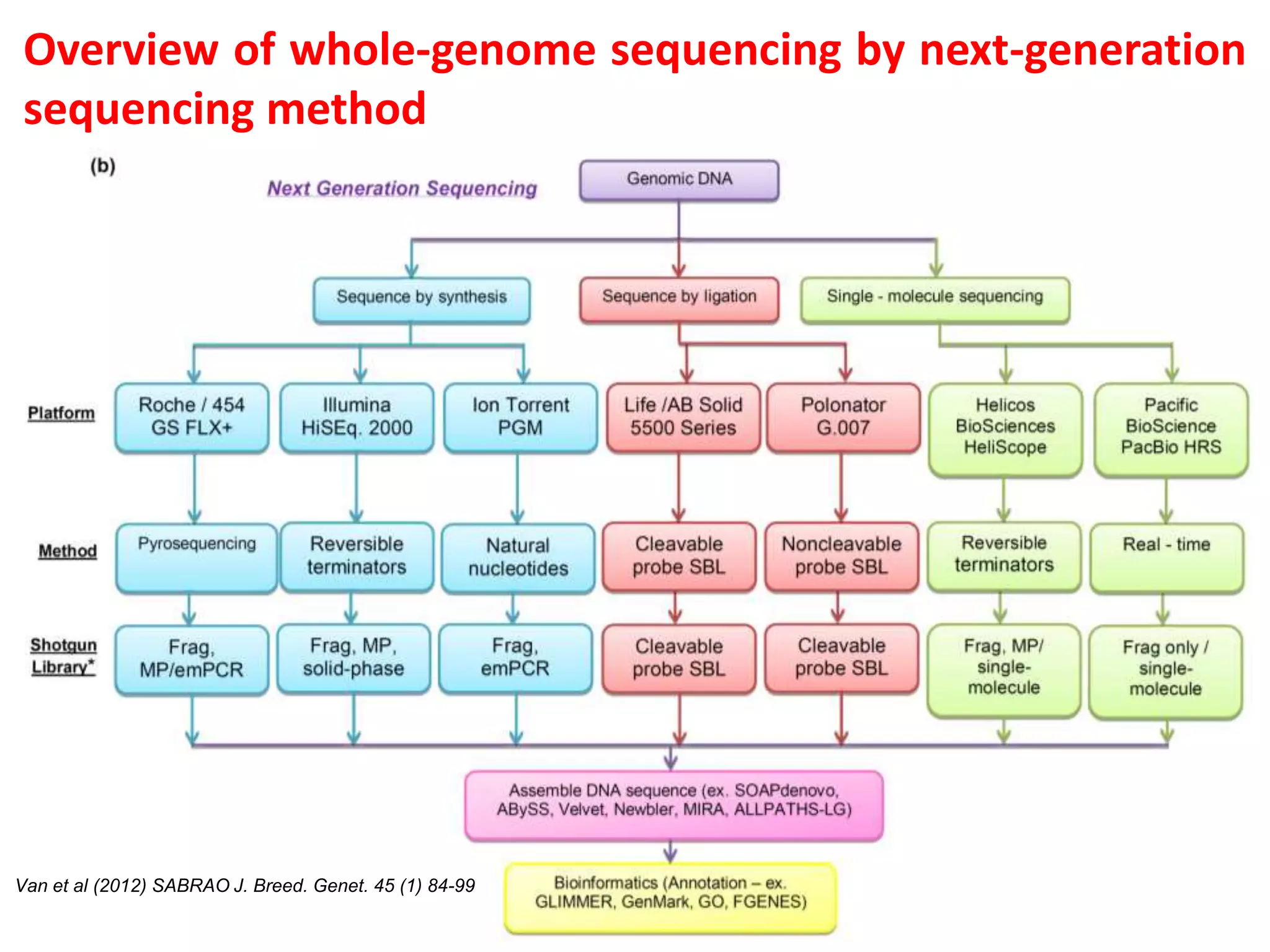
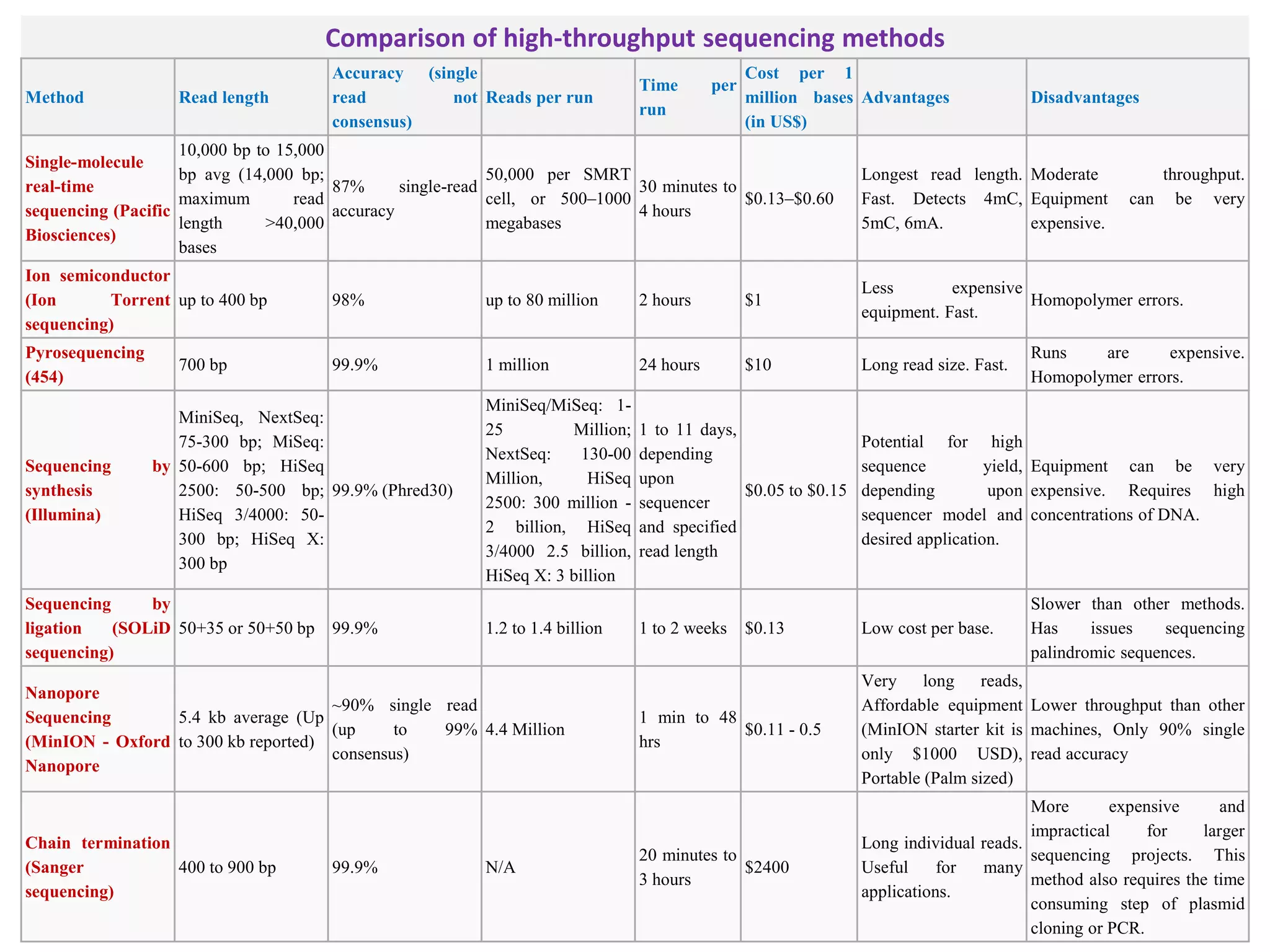
The document discusses various DNA and RNA sequencing methods and technologies. It begins with an overview of sequencing-based markers like DNA sequencing, RNA sequencing, SNPs, epigenetic markers, and omics. The document then provides more details on the history and development of sequencing technologies, including early methods like Sanger and Maxam-Gilbert sequencing. It discusses next generation sequencing platforms like MPSS, 454 pyrosequencing, Illumina, Ion Torrent, ABI-SOLiD, and their approaches. The document concludes with an overview of third generation long-read sequencing technologies like SMRT and nanopore sequencing.