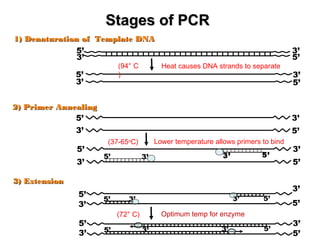
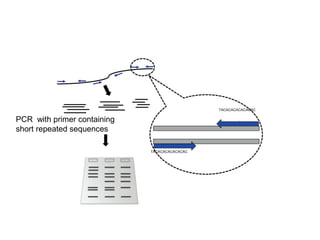
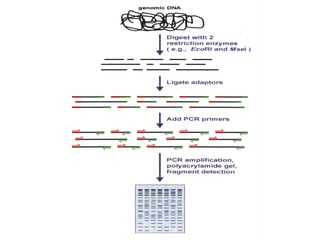
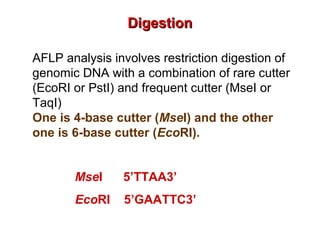
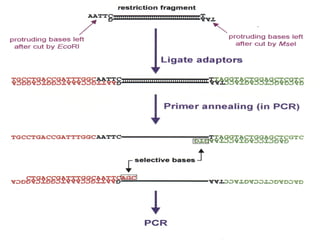
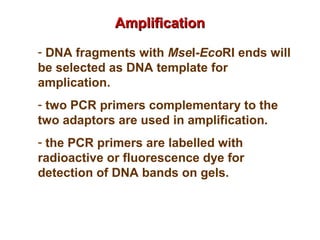
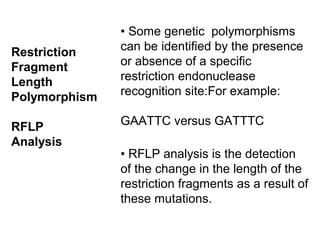
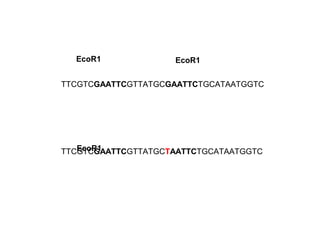
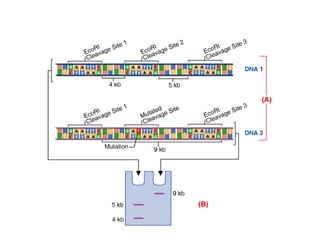
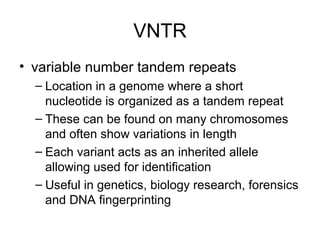
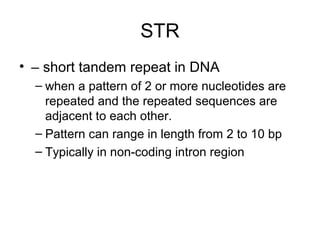
The document discusses various types of genetic markers that can be used to measure genetic diversity, including random amplified polymorphic DNA (RAPD), inter-simple sequence repeats (ISSR), amplified fragment length polymorphism (AFLP), restriction fragment length polymorphism (RFLP), microsatellites, minisatellites, and mitochondrial DNA markers. It provides details on how each type of marker works and its applications in studying genetic variation, relationships, and evolution.