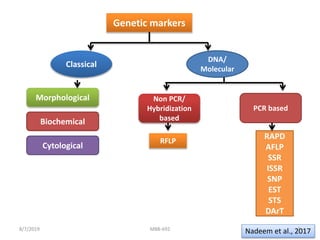
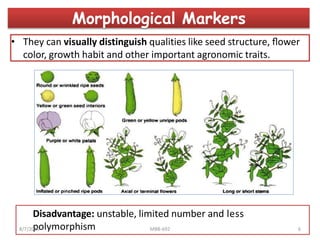
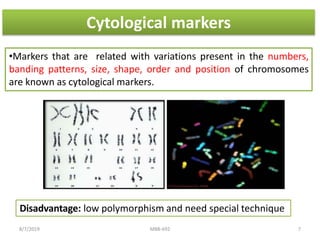
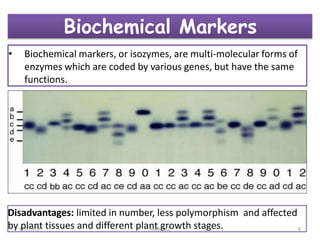
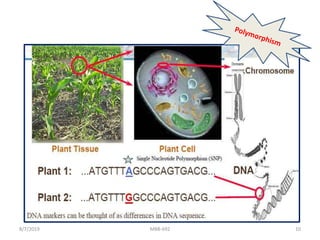
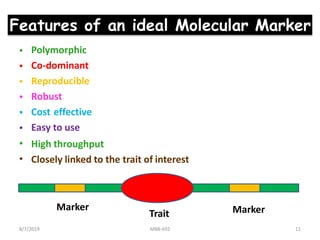
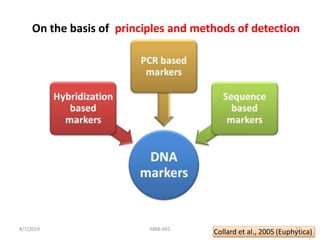
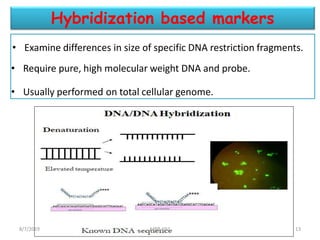
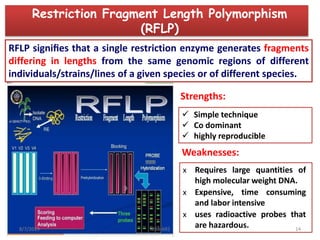
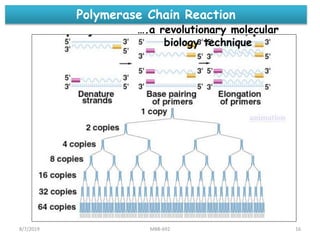
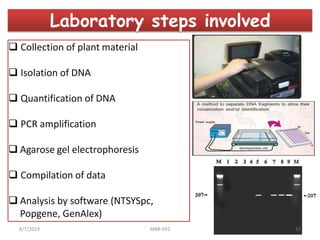
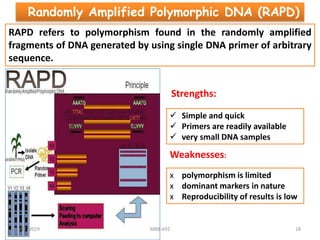
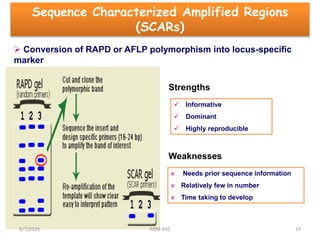
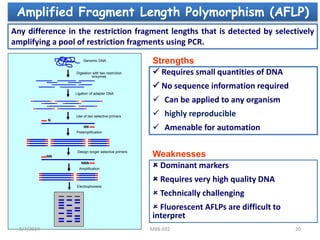
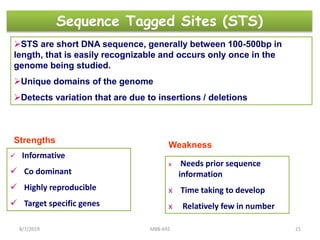
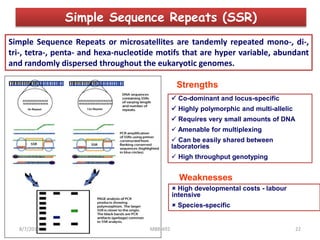
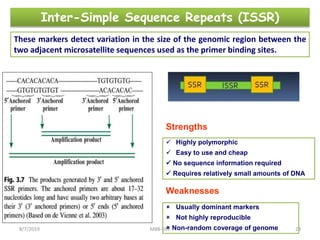
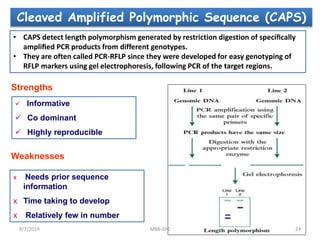
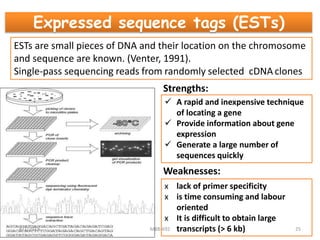
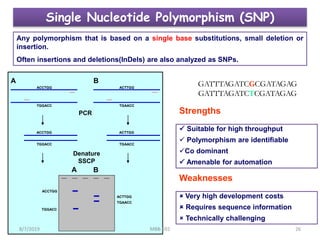
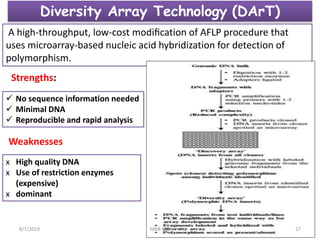
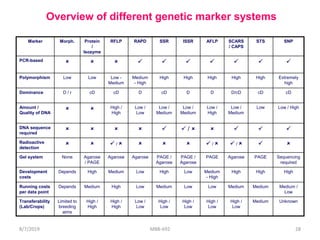
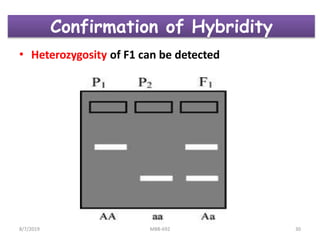
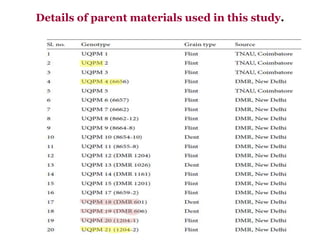
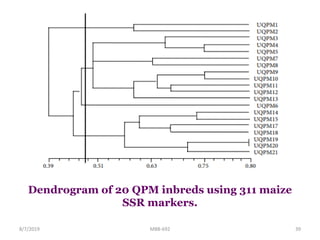
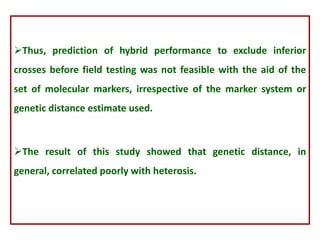
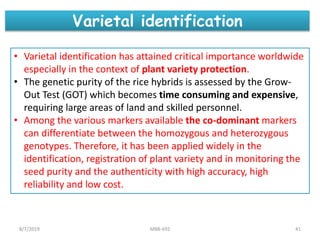
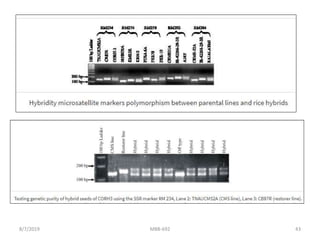
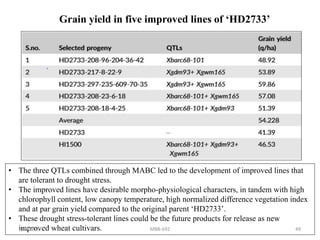
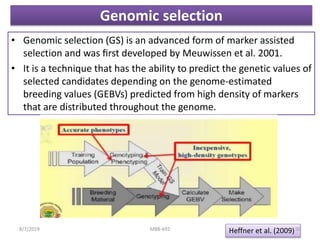
This document provides an overview of molecular markers that can be used for crop improvement. It discusses different types of markers such as morphological, cytological, biochemical, DNA-based markers. DNA-based markers are further classified into hybridization-based markers like RFLP and PCR-based markers like RAPD, AFLP, SSR, ISSR, SNP. The document compares various marker techniques and provides their principles, strengths, weaknesses and applications in crop breeding programs. Molecular markers can be useful for tasks like hybrid purity testing, genetic diversity analysis, linkage mapping and marker-assisted selection.