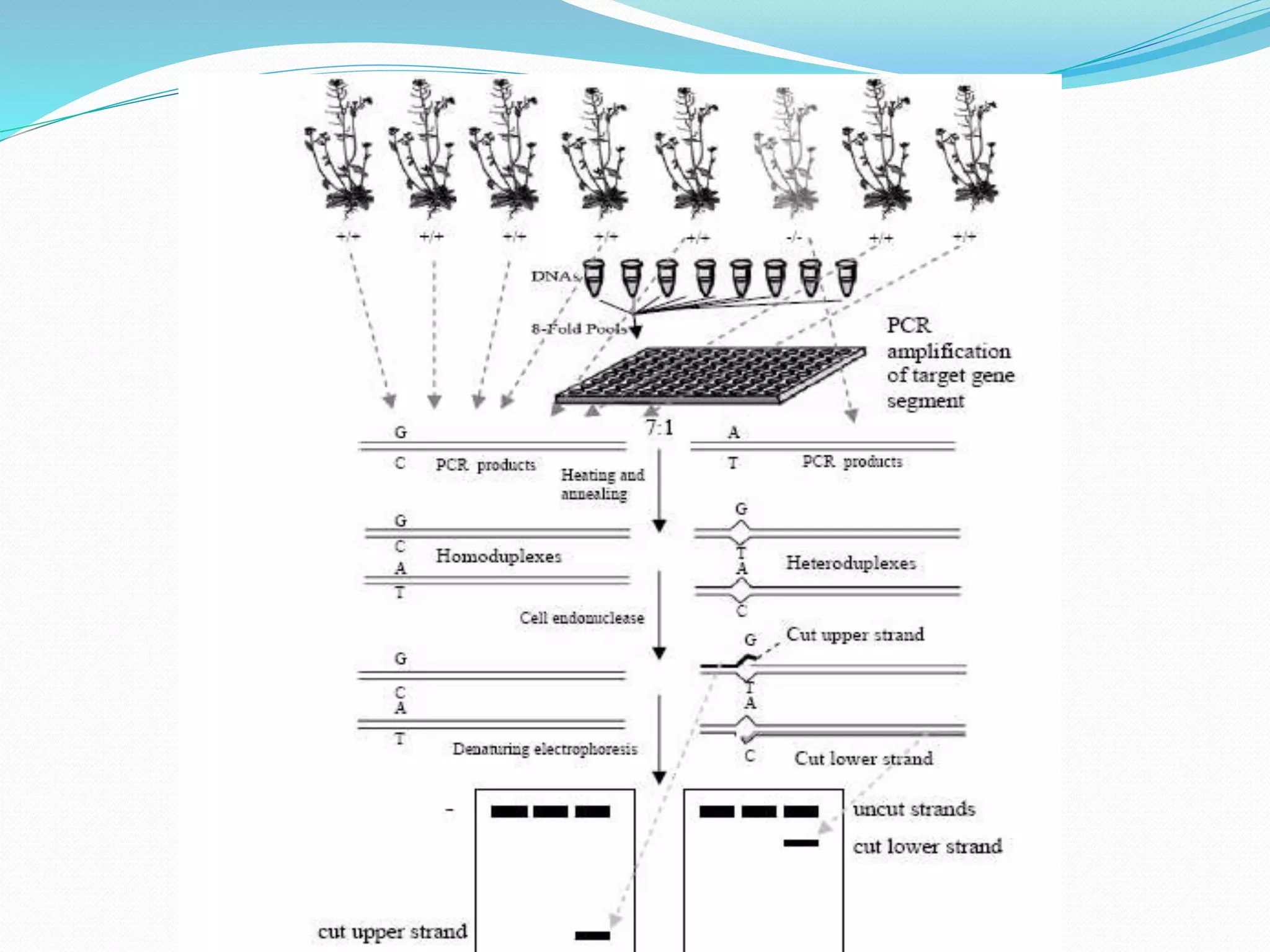
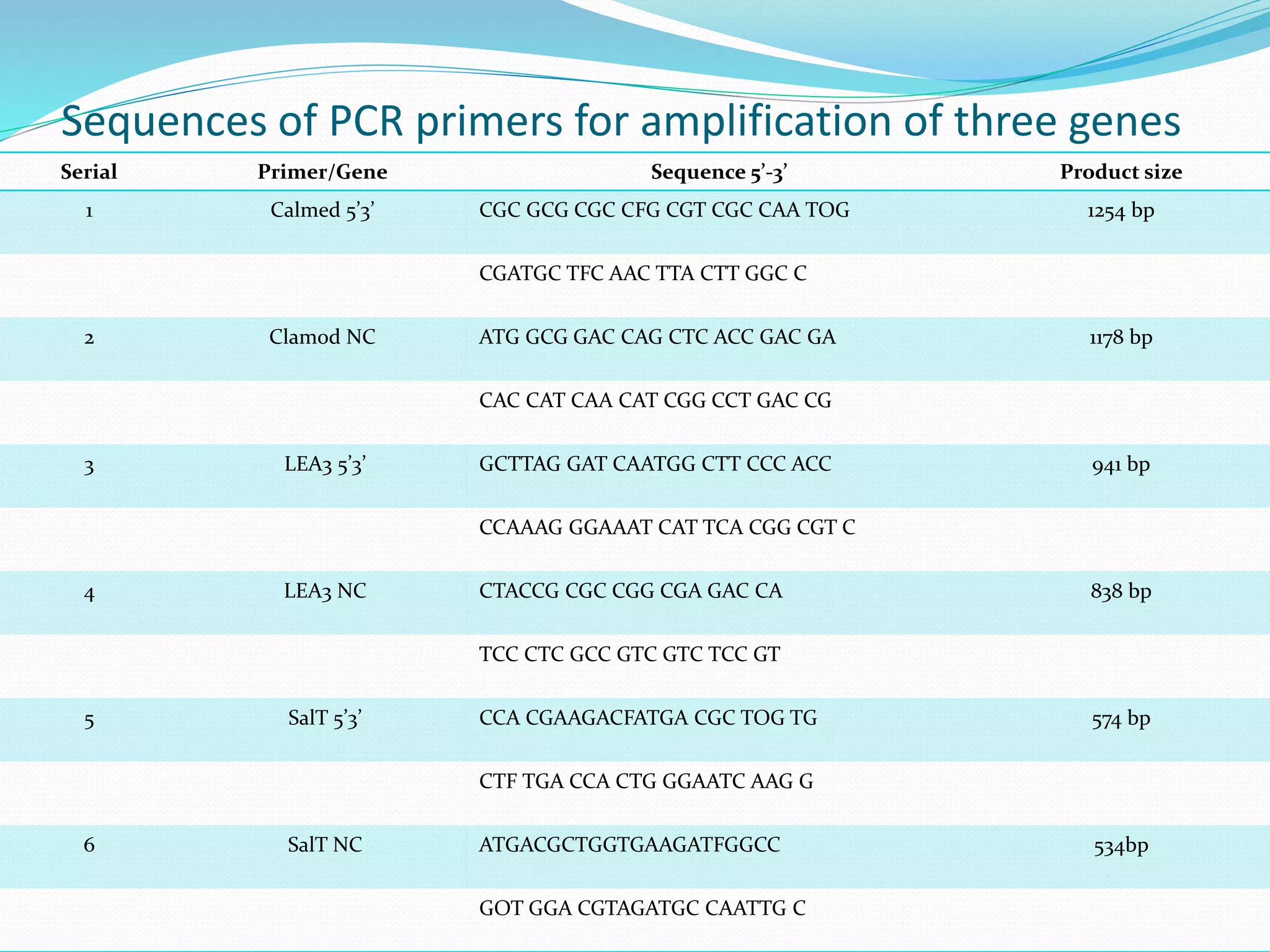
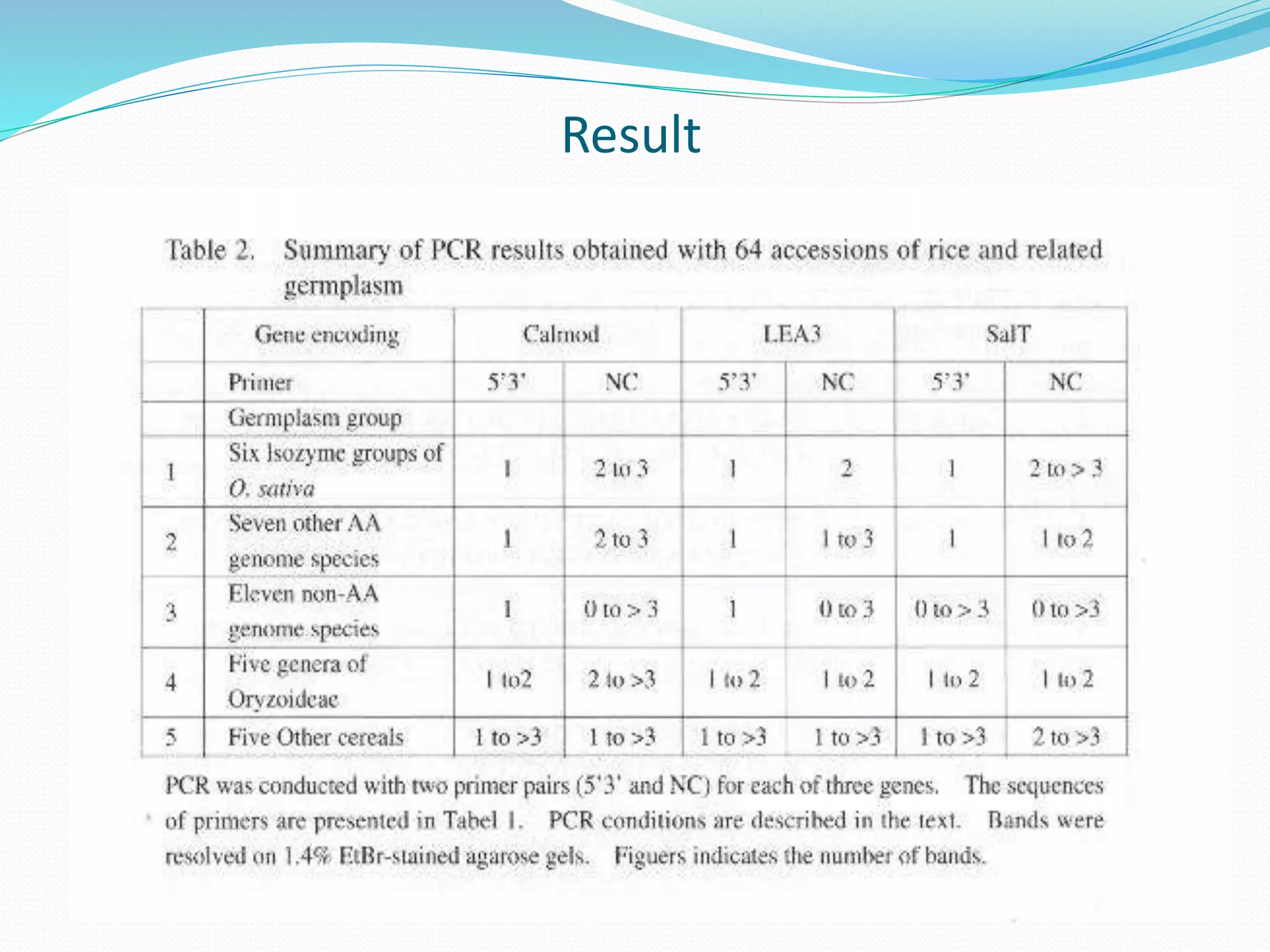
The document discusses allele mining, which aims to identify allelic variations in genetic resources collections that are relevant for traits of interest. It describes how allele mining works to unlock hidden genetic variation by identifying single nucleotide polymorphisms and new haplotypes. The document then provides details on a case study of allele mining focused on three genes - calmodulin, LEA3, and SalT - important for abiotic stress tolerance in rice and related species. Primers were developed to amplify regions of these three genes from 64 accessions representing rice and other grasses.