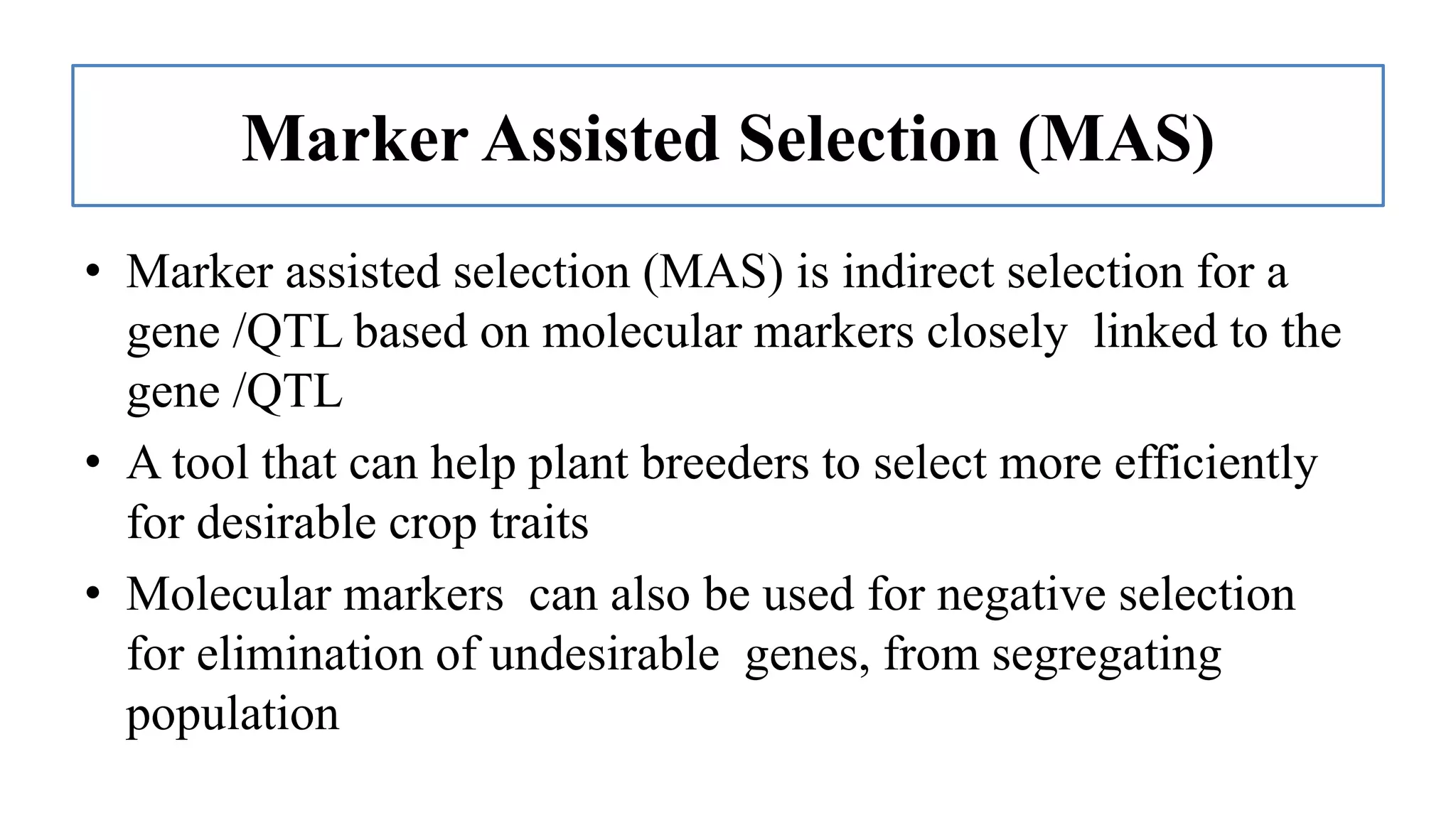
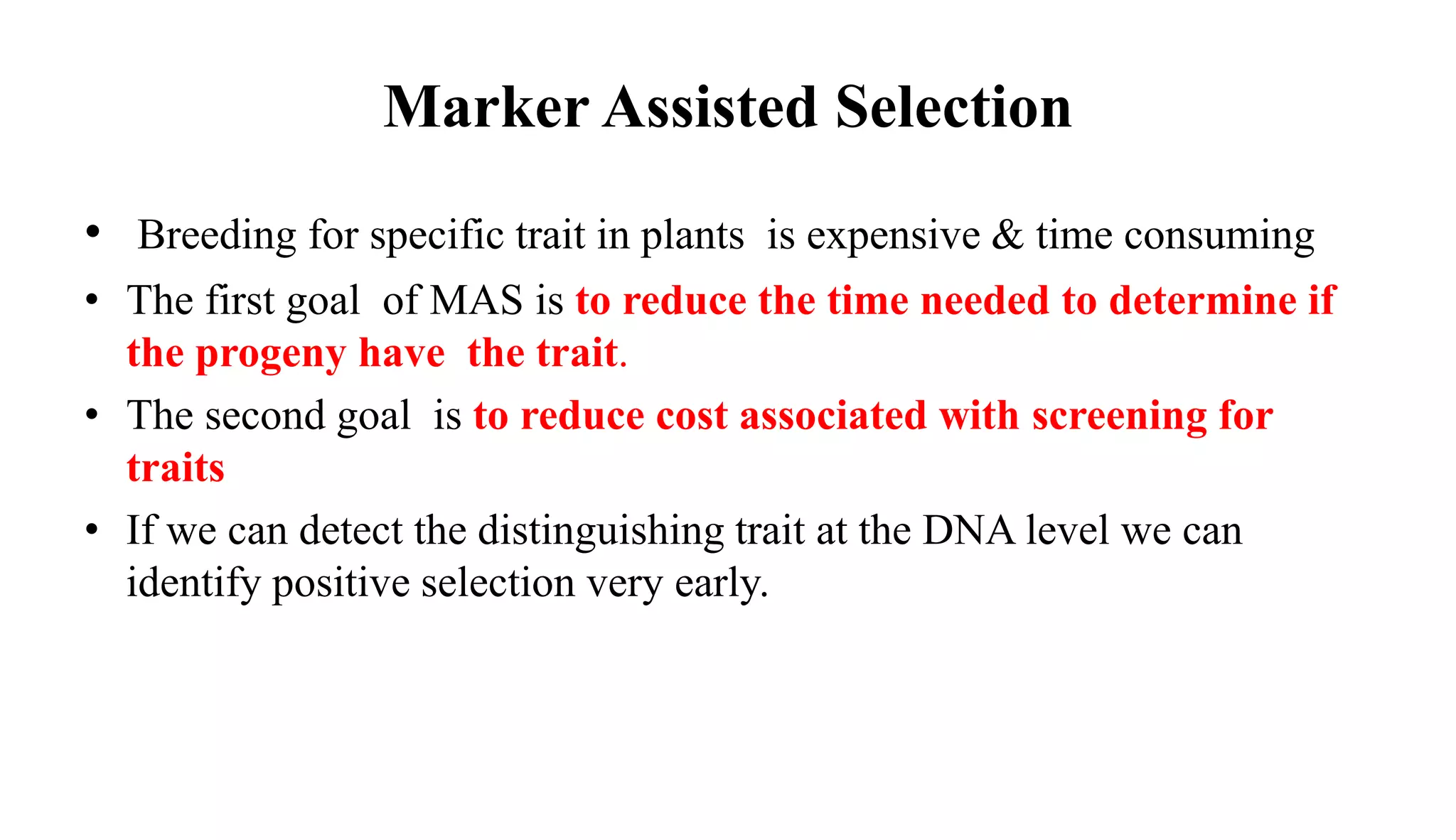
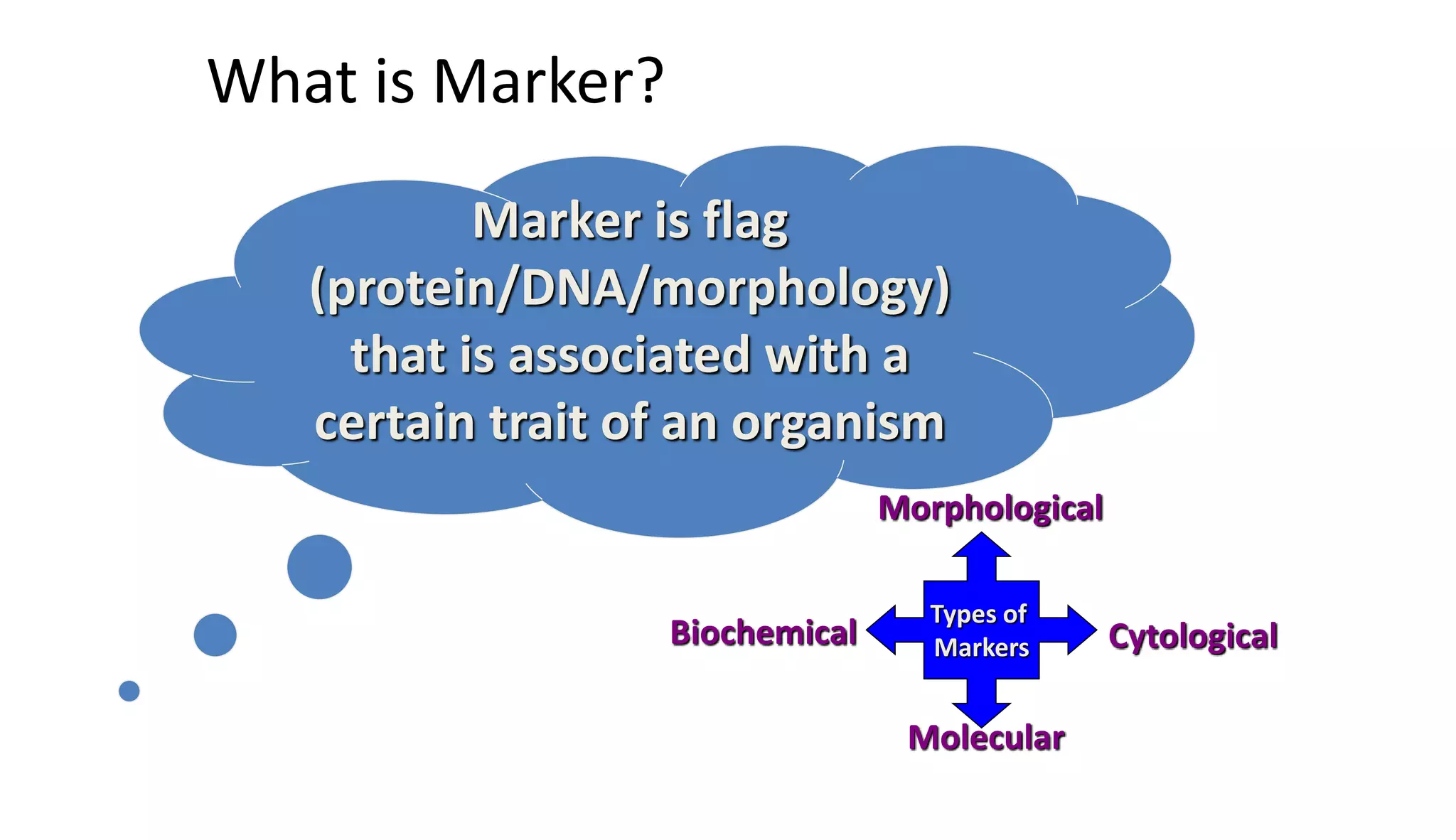
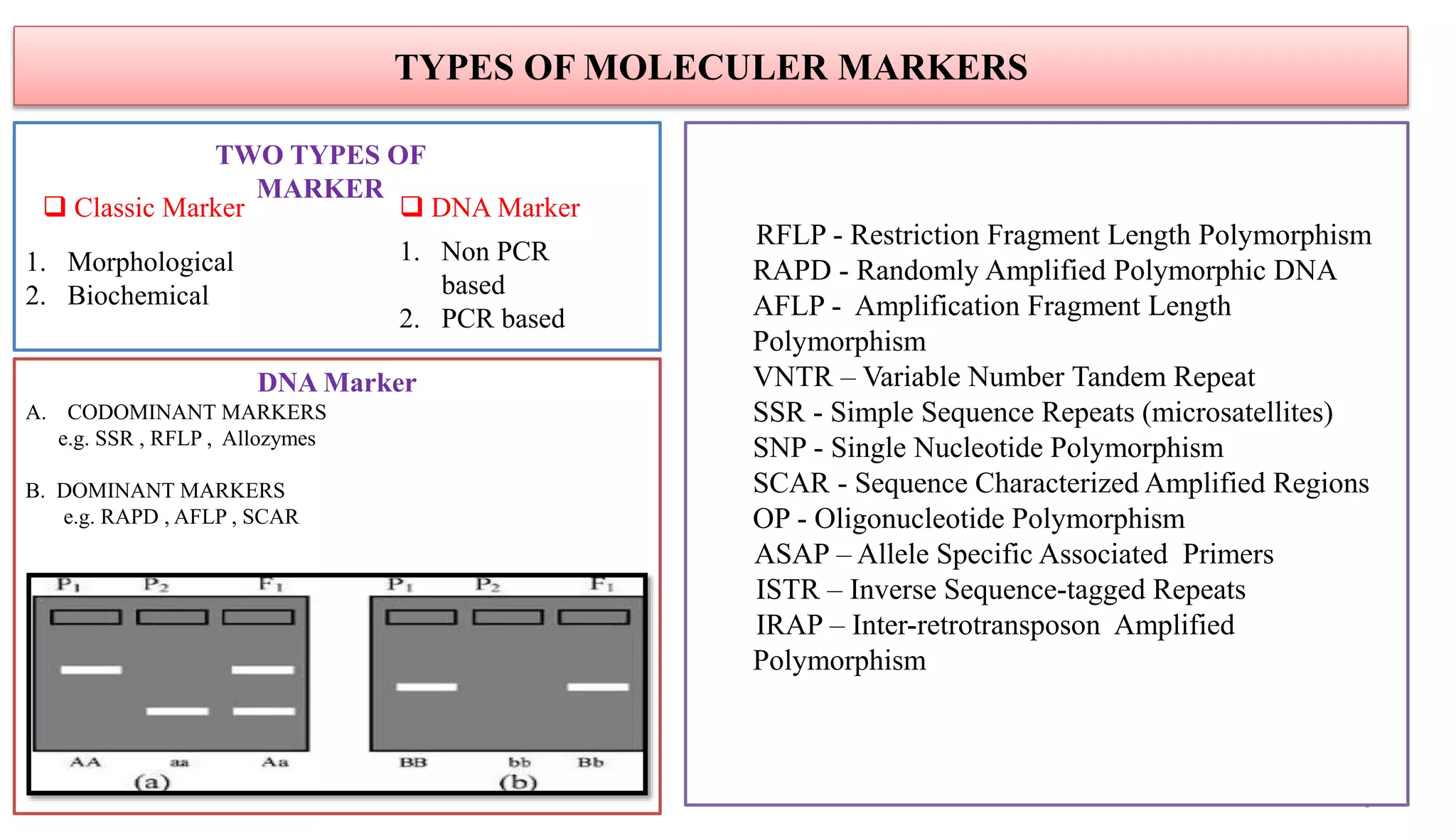
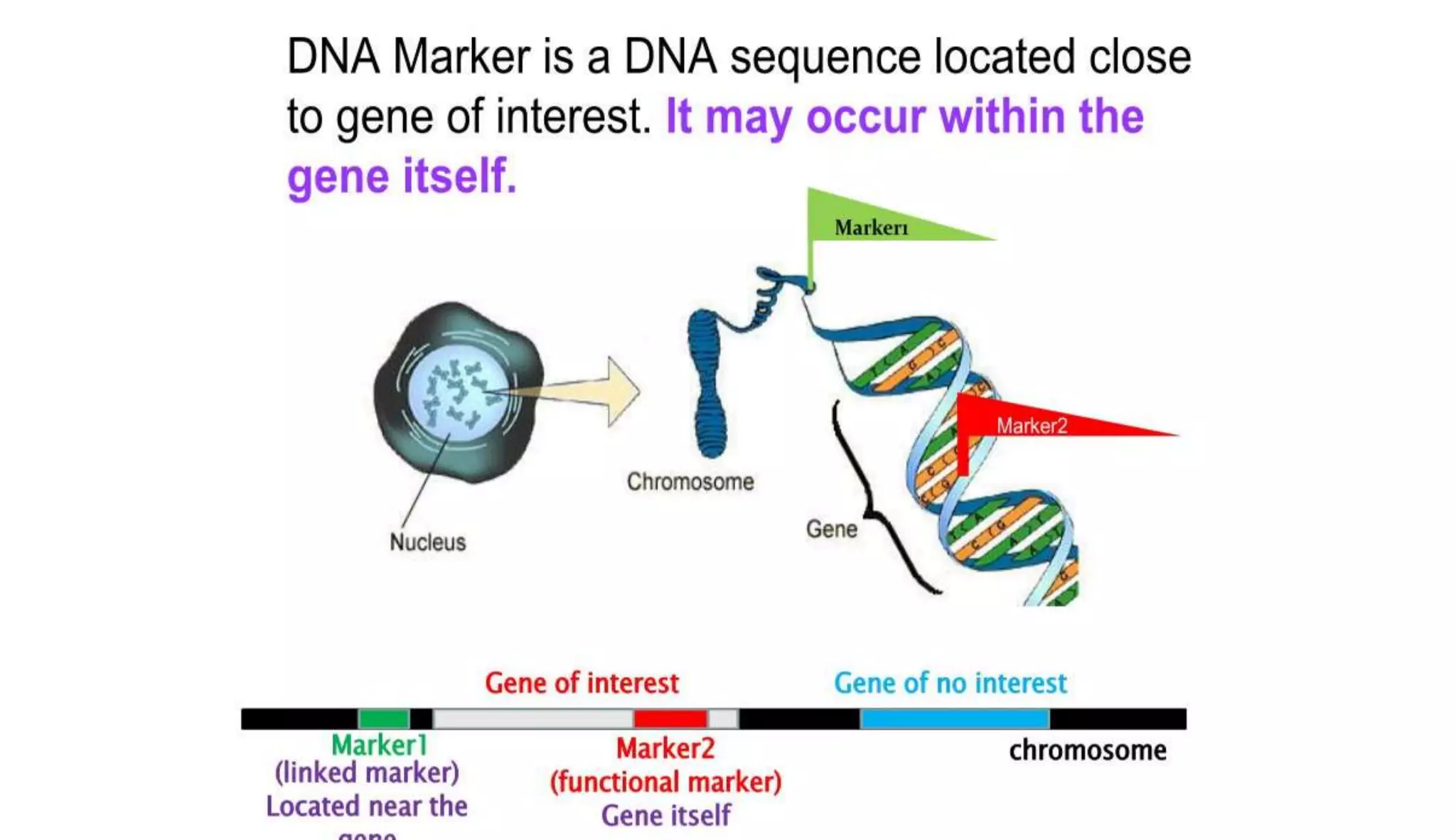
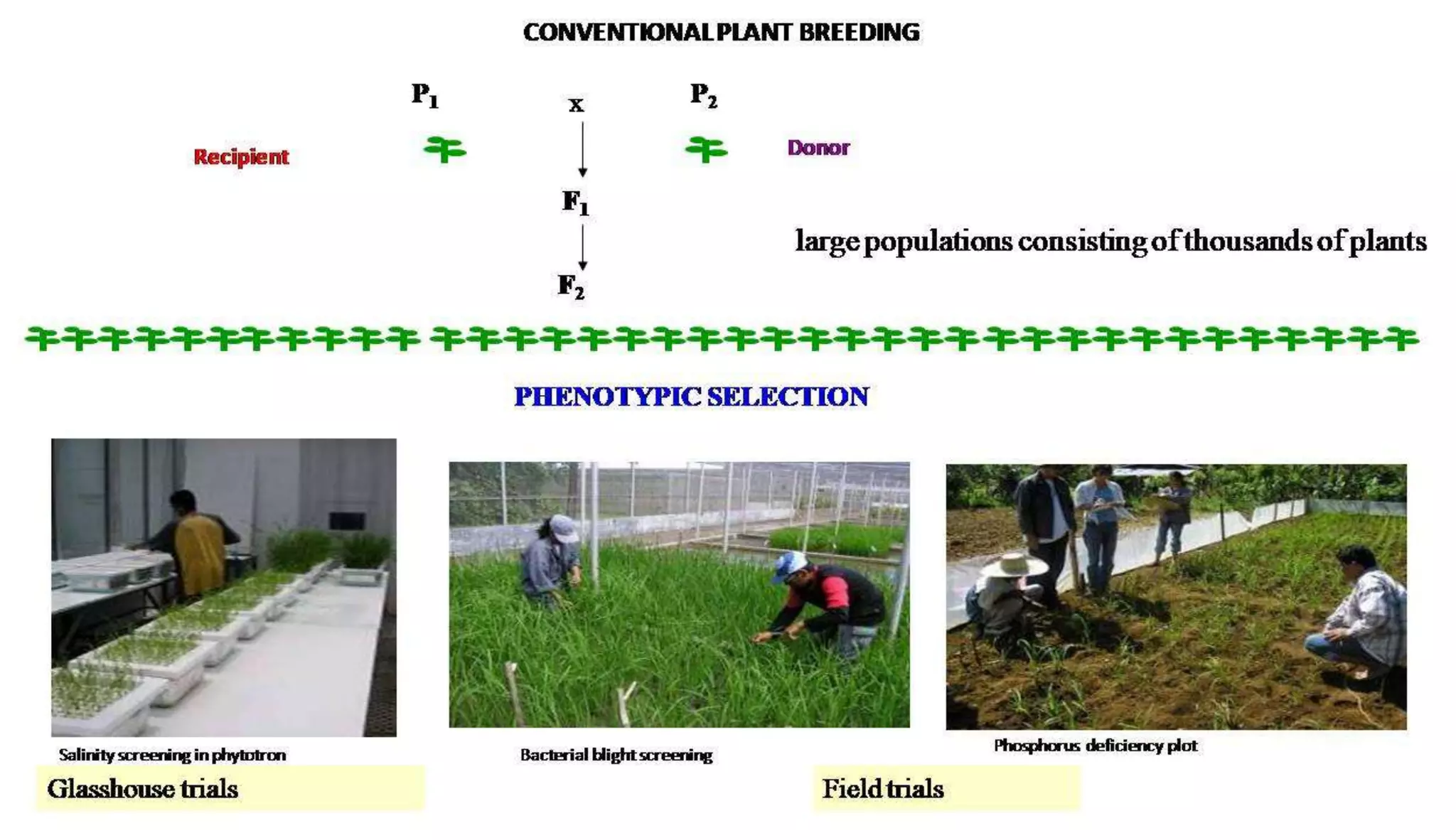
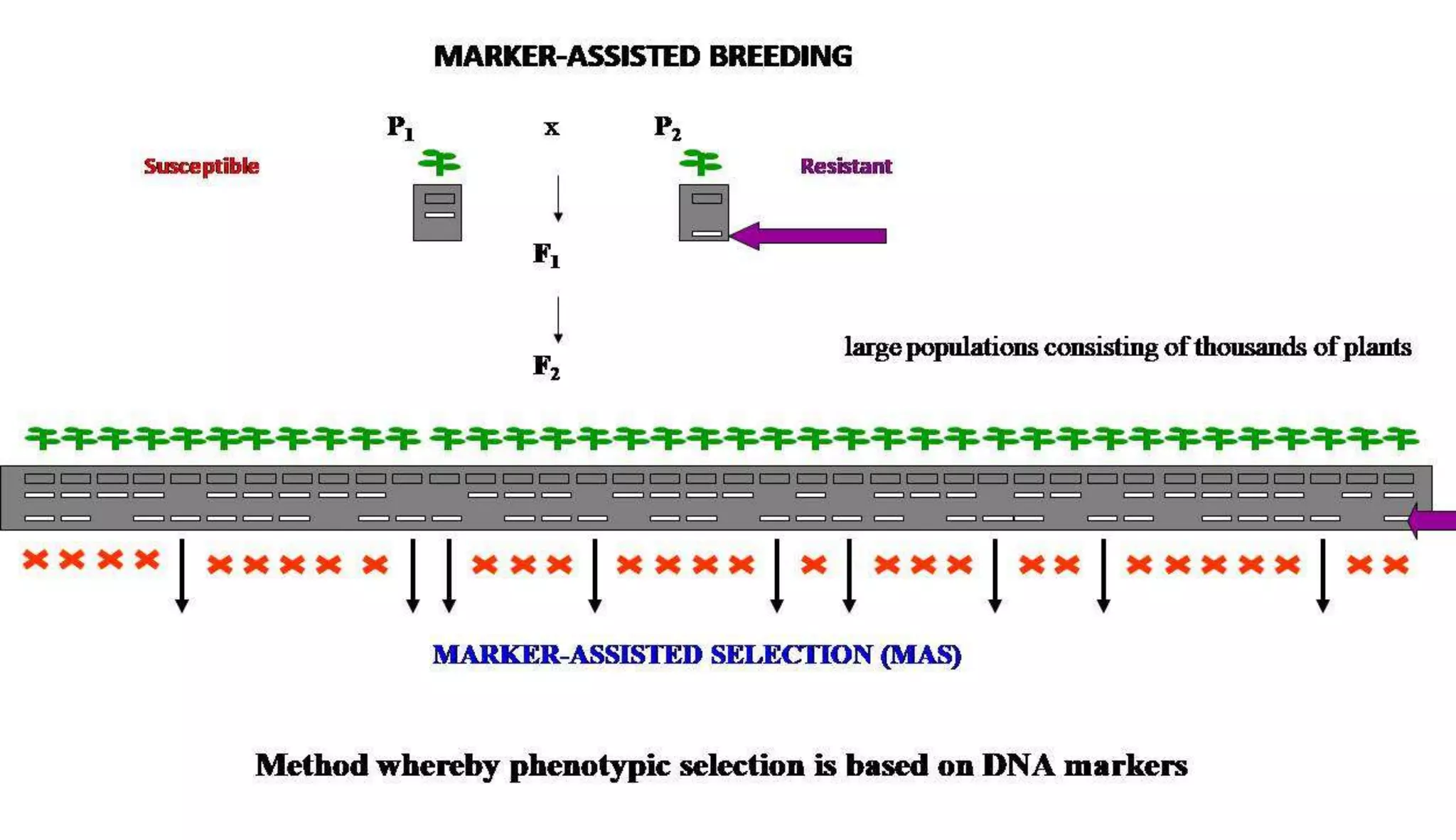
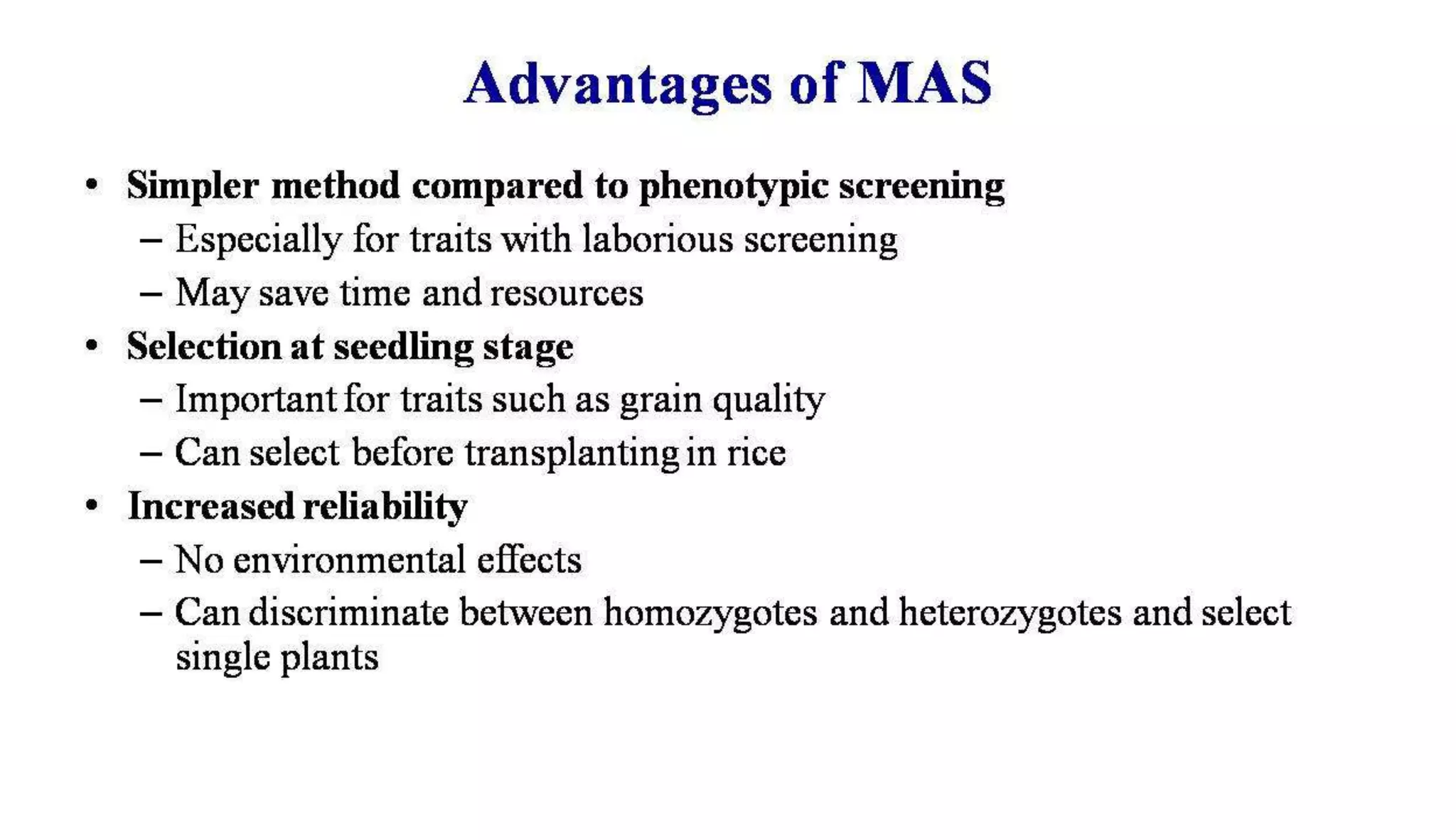
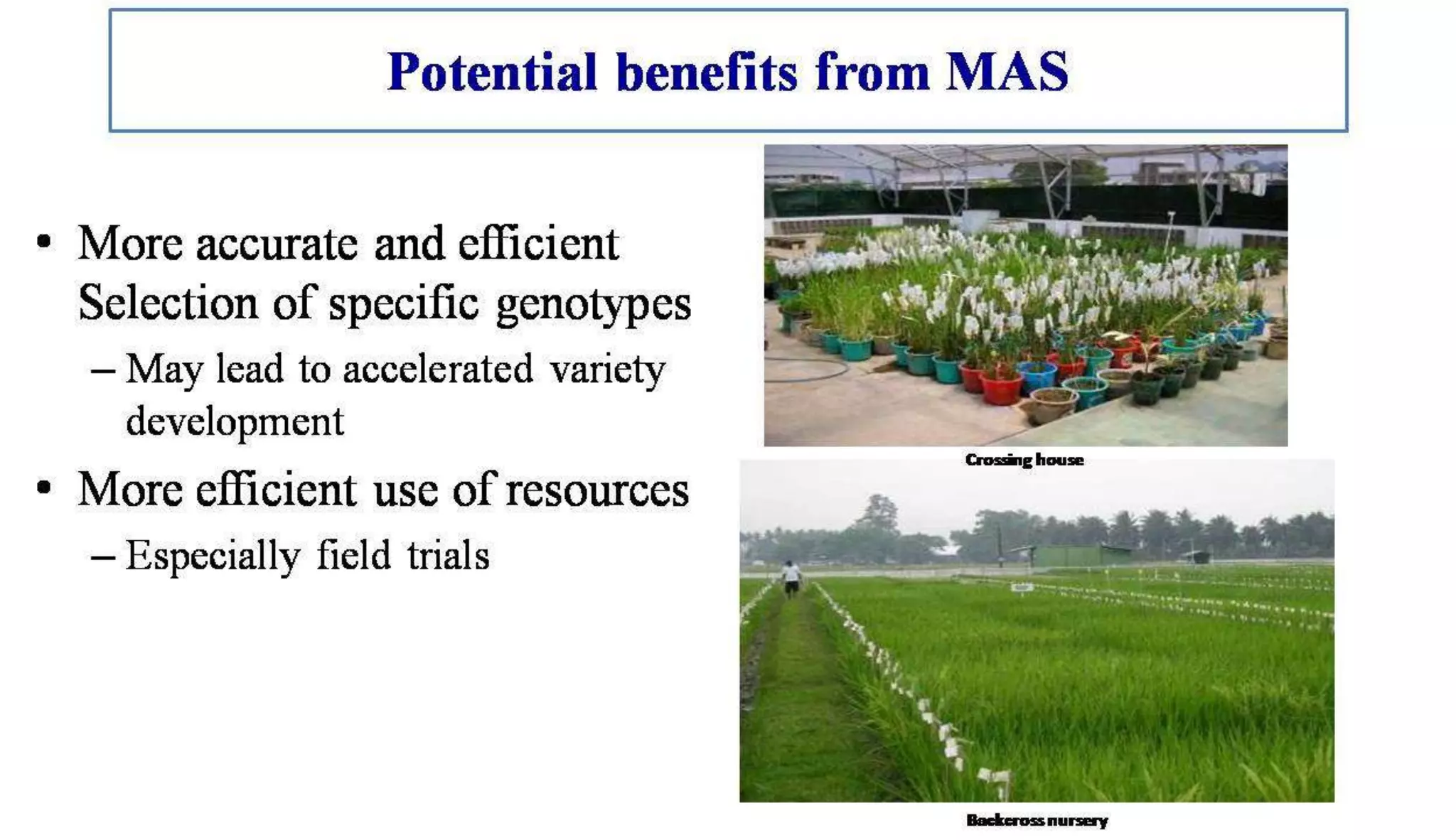
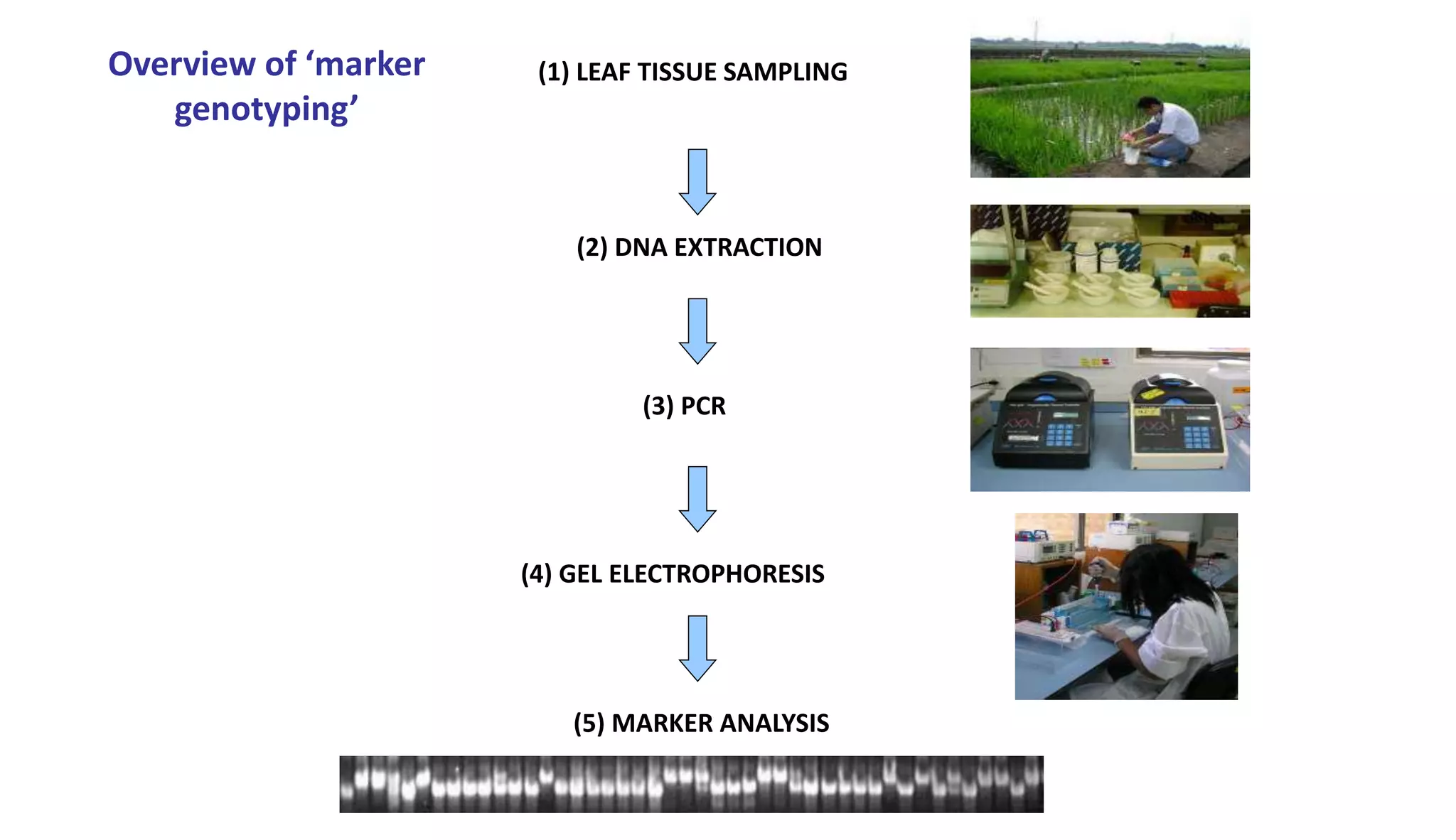
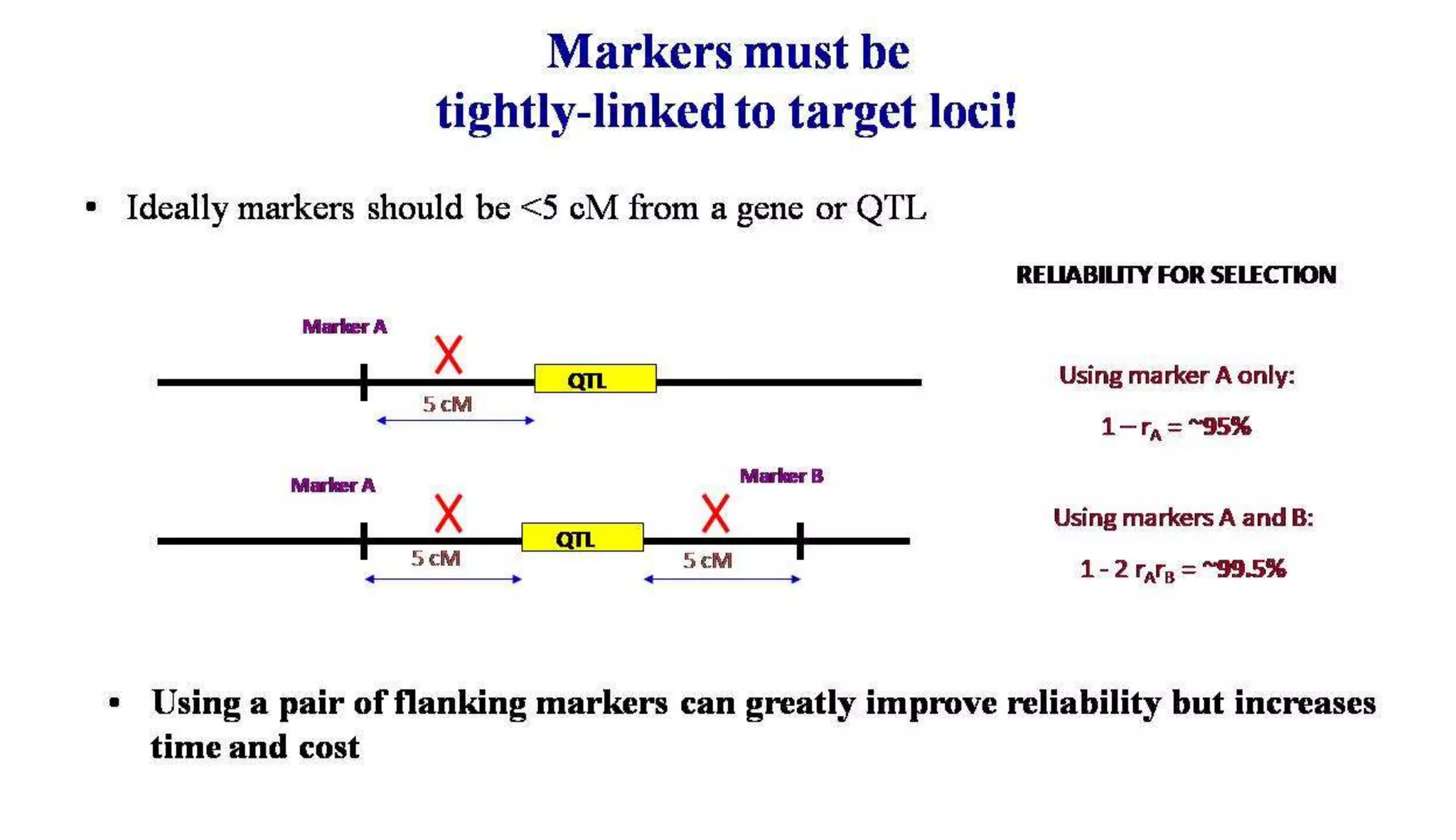
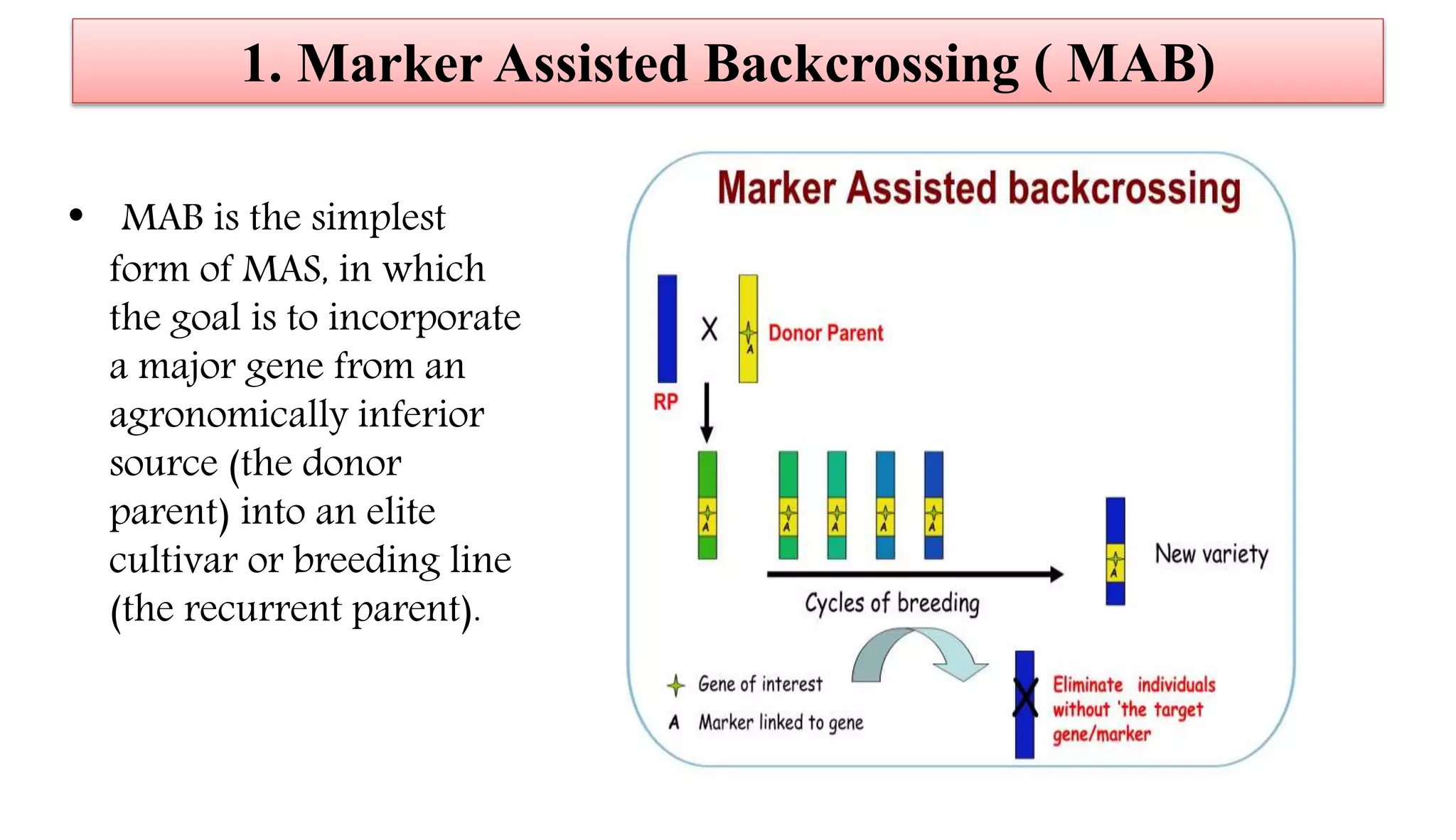
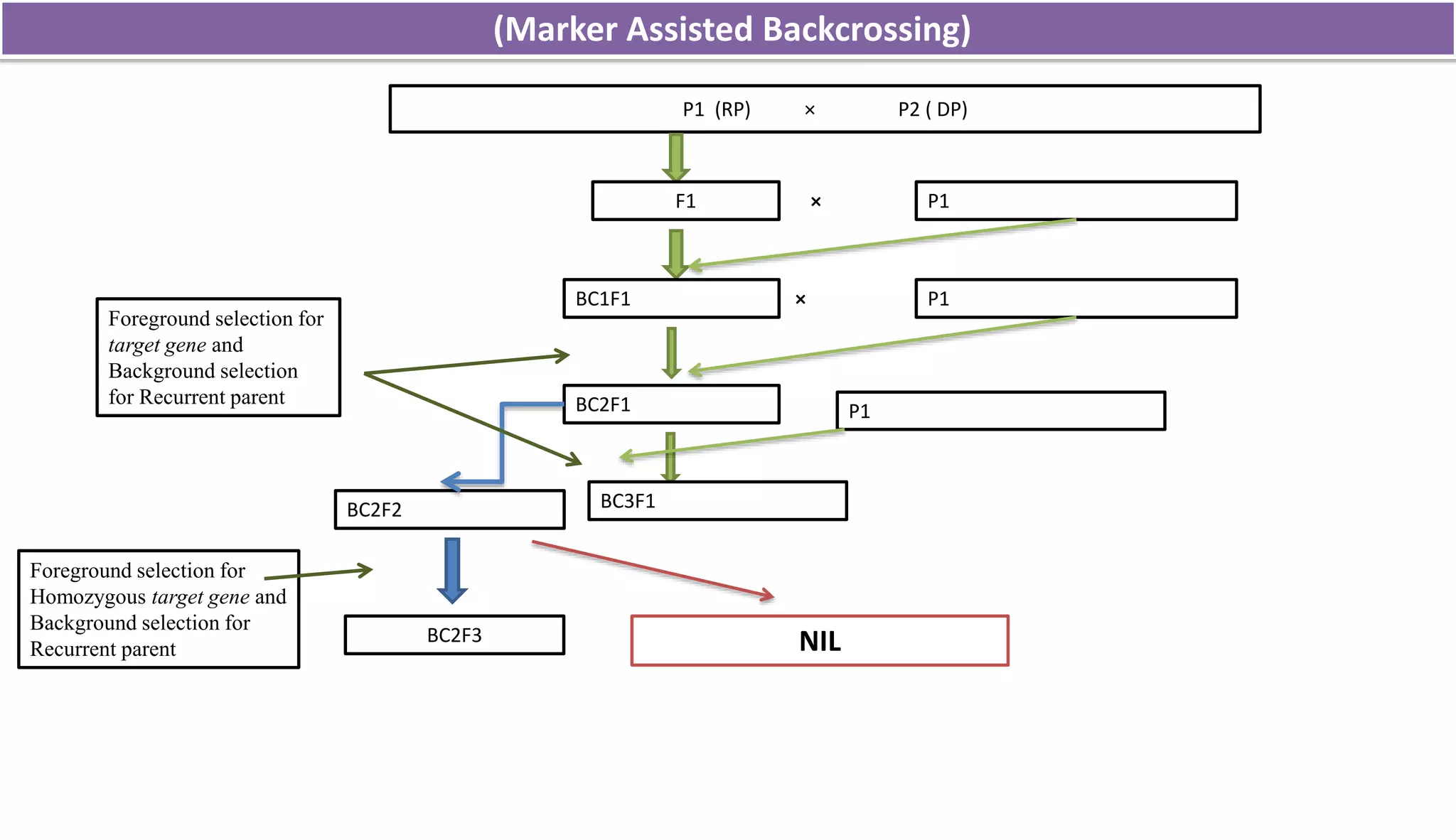
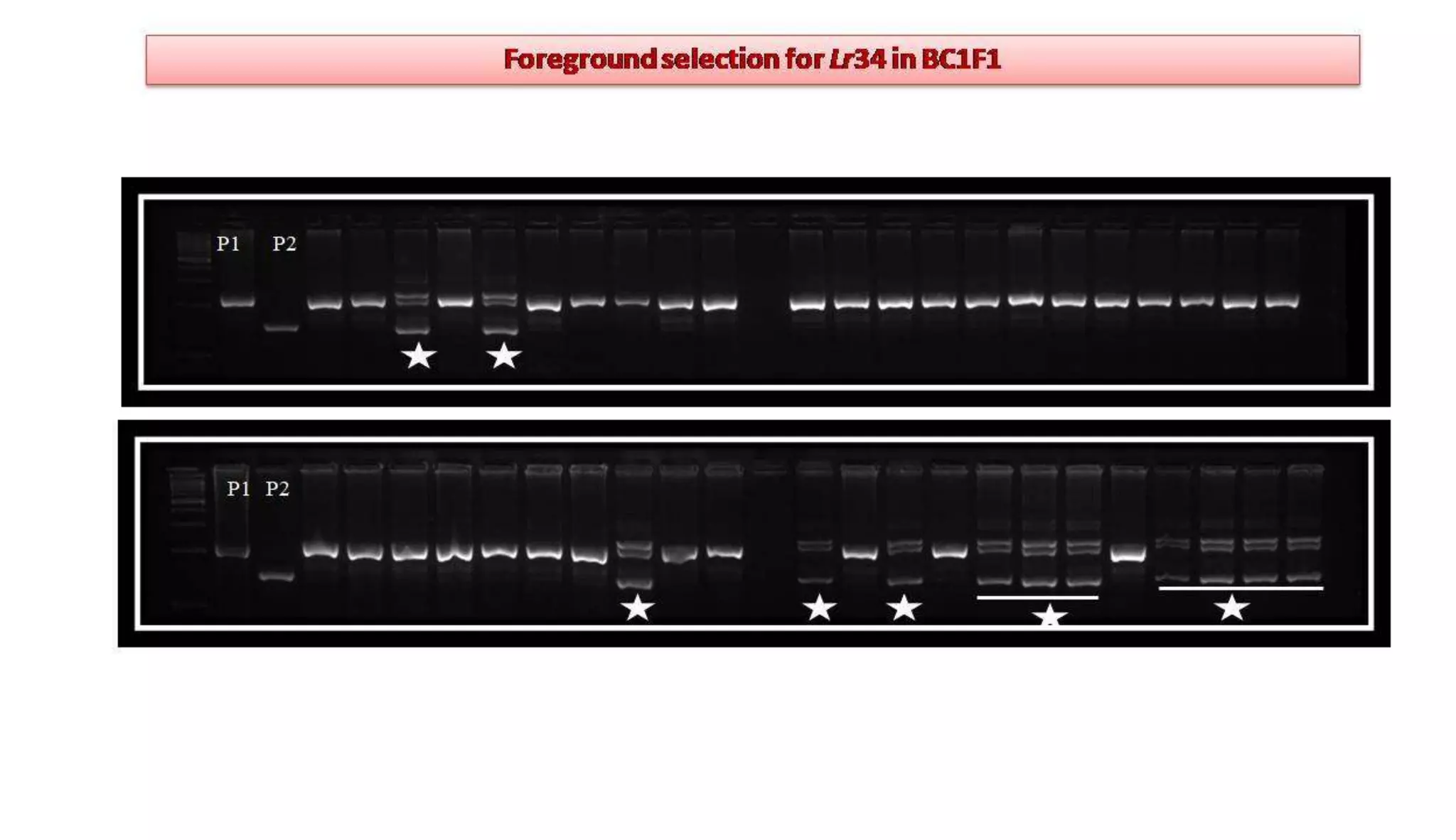
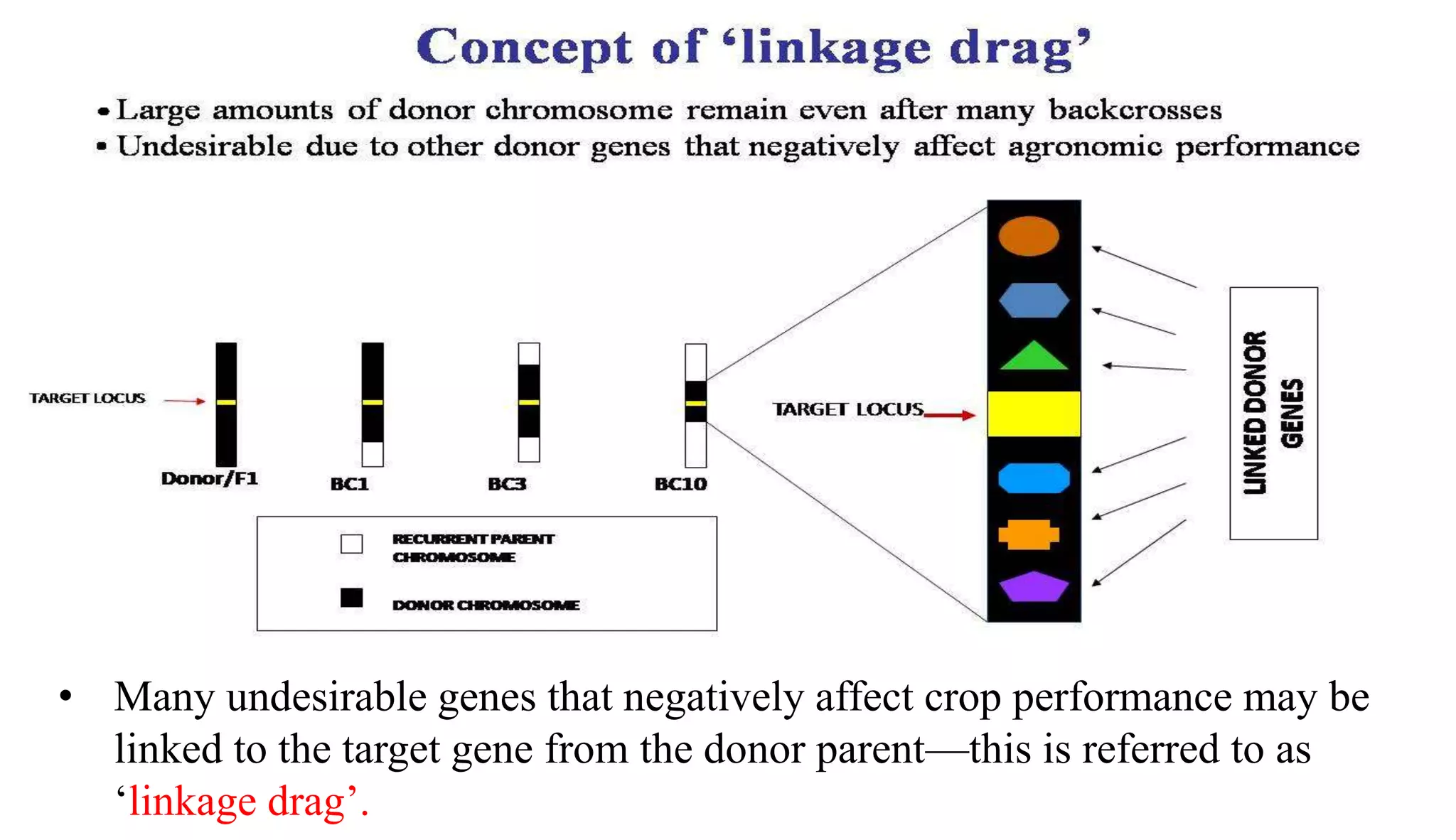
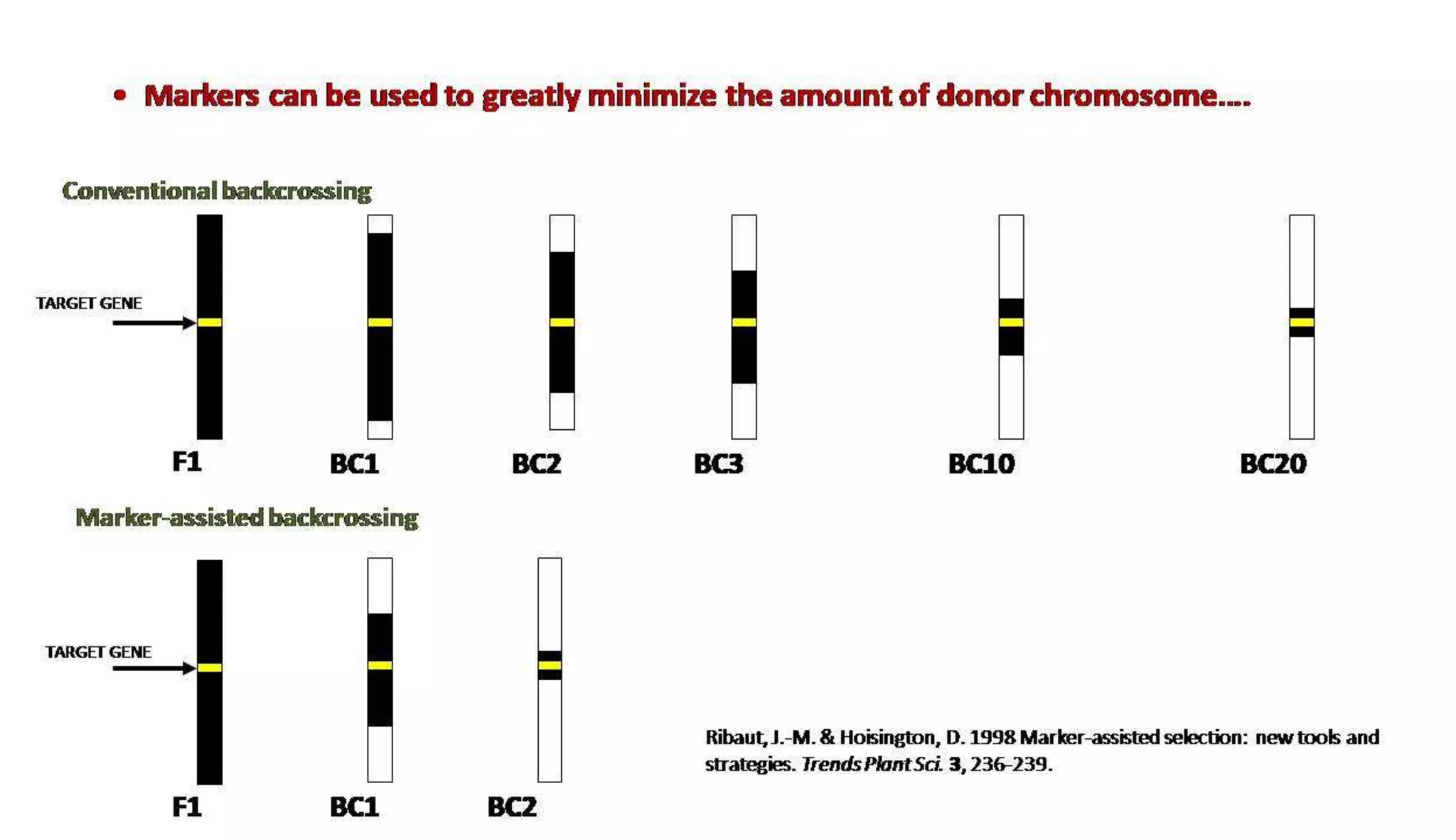
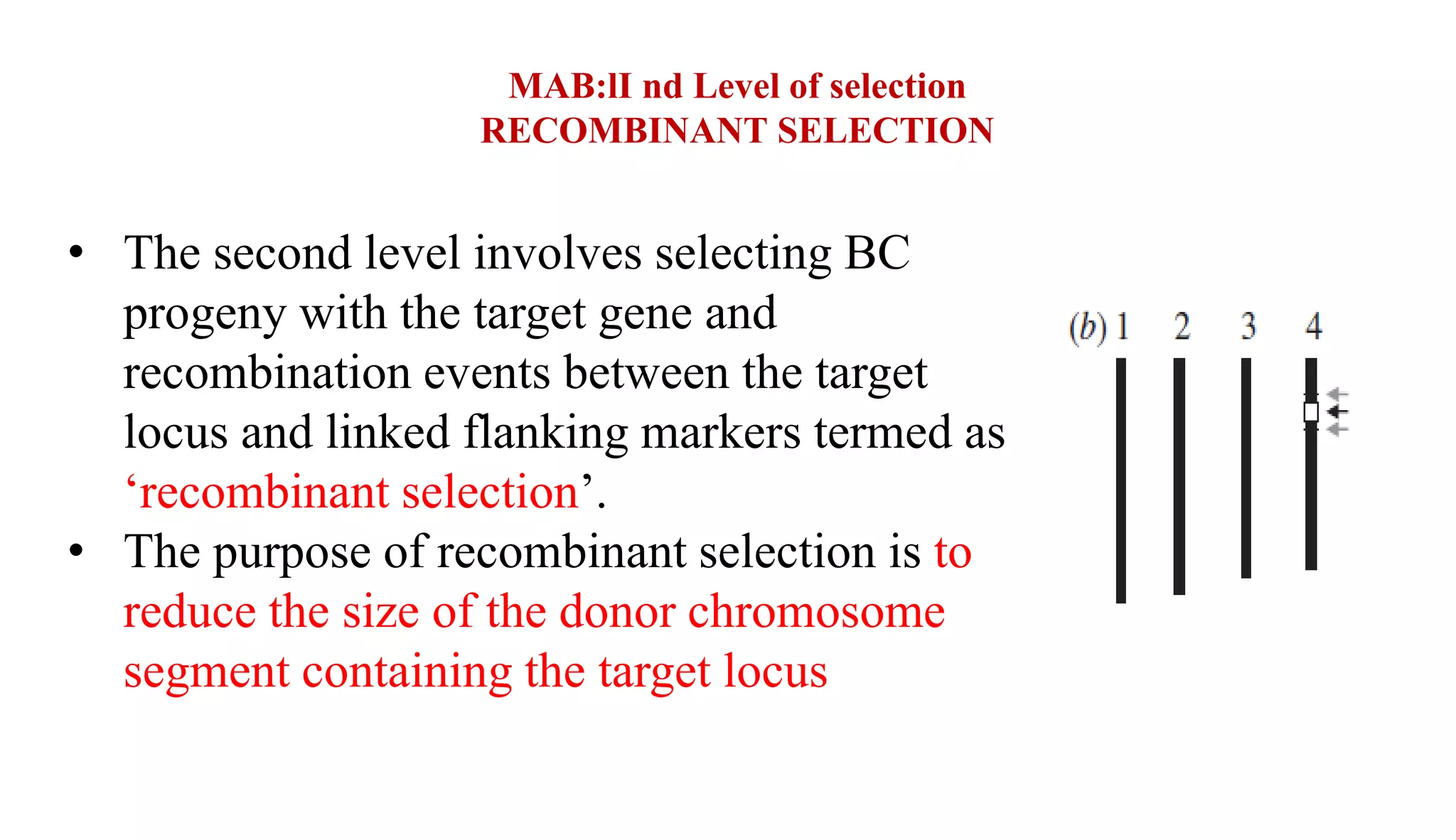
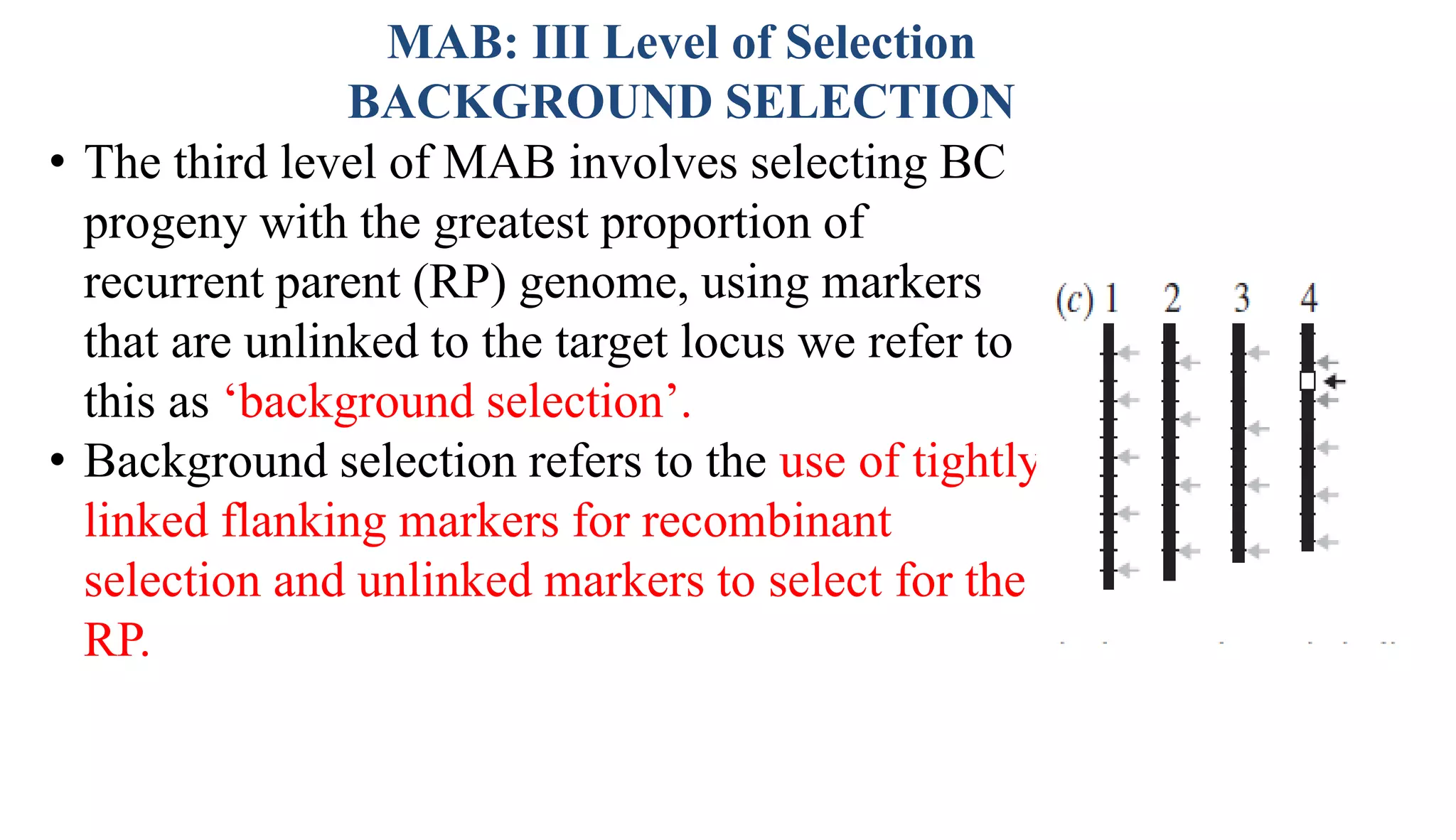
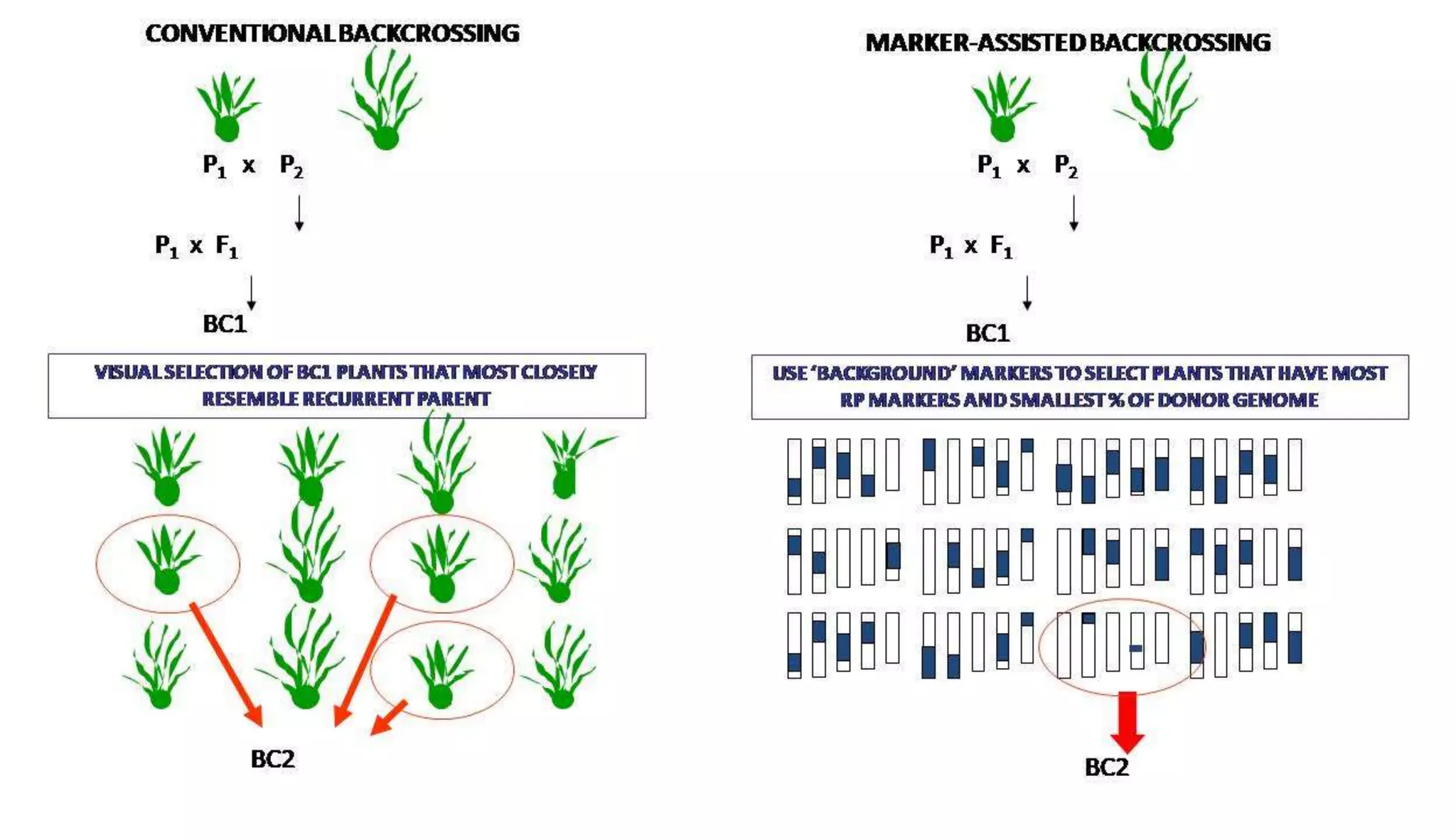
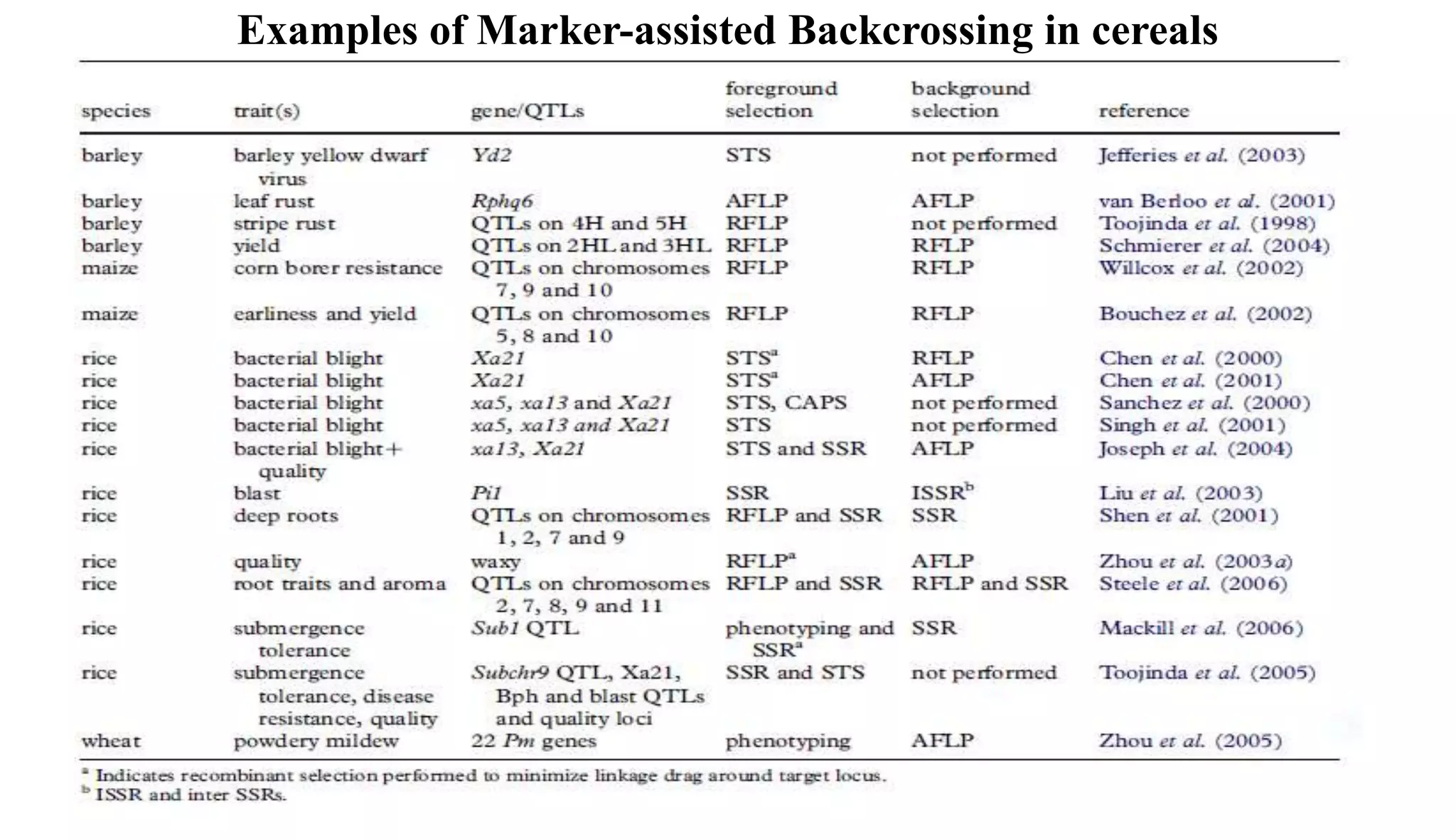
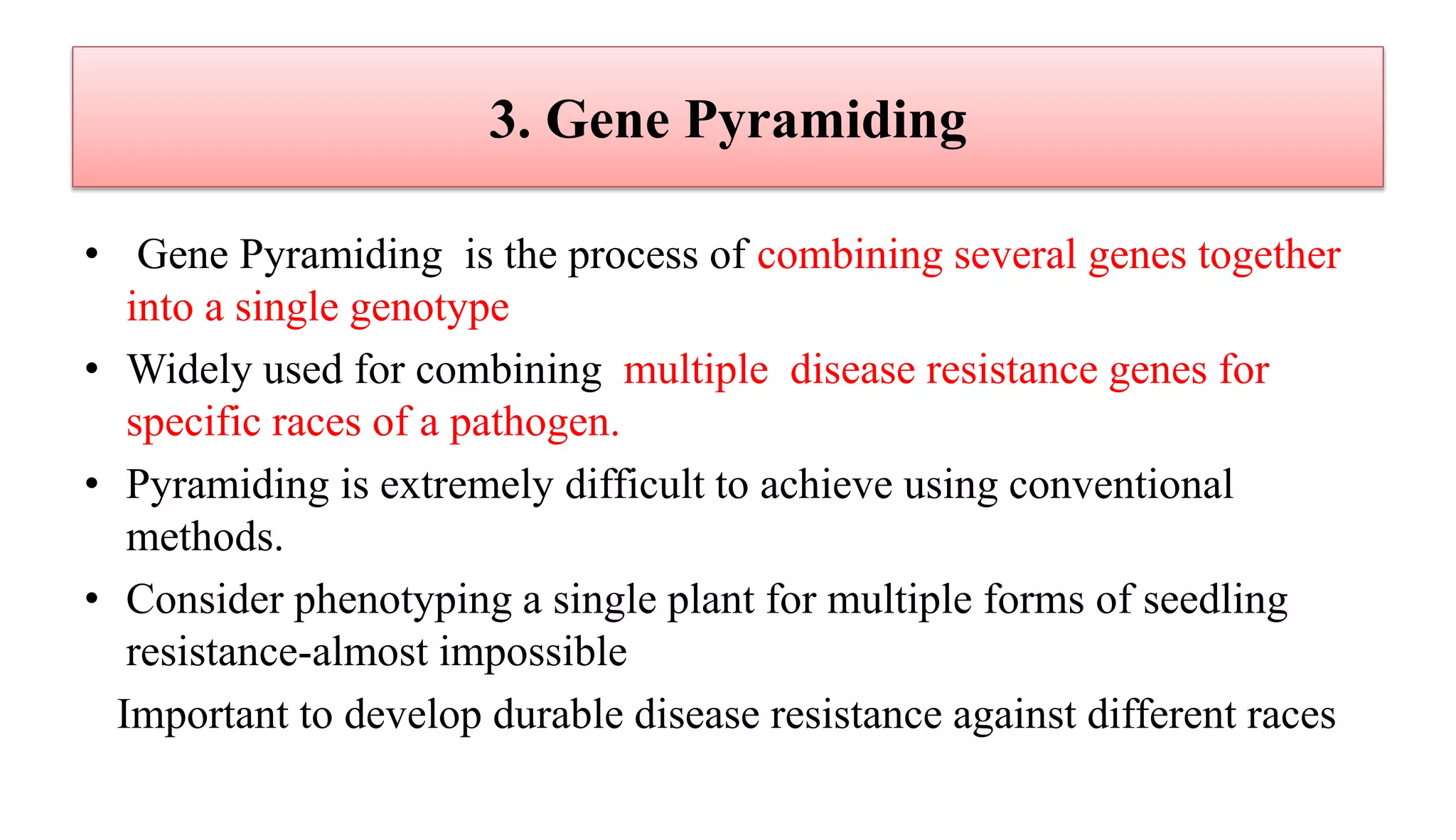
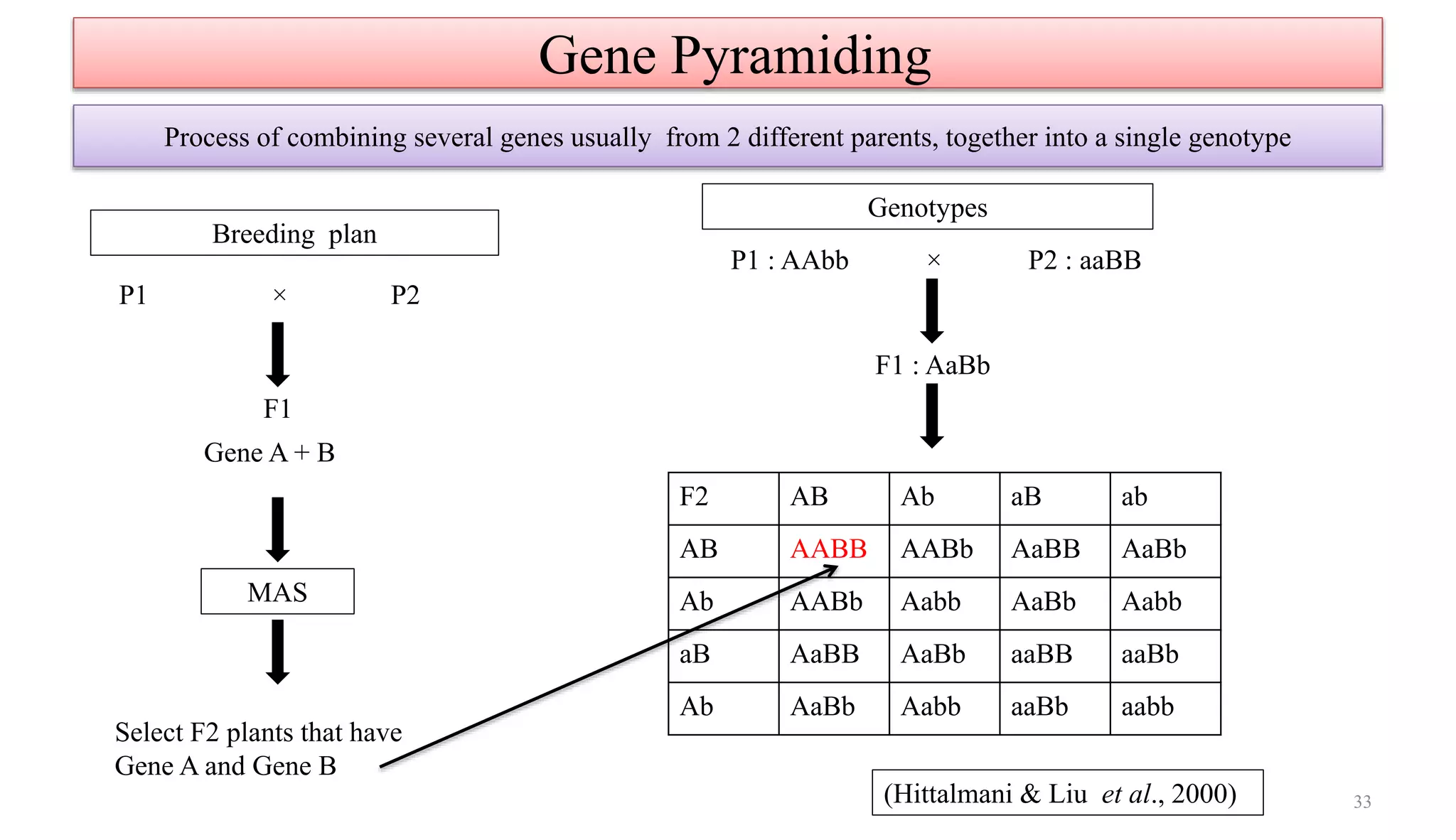
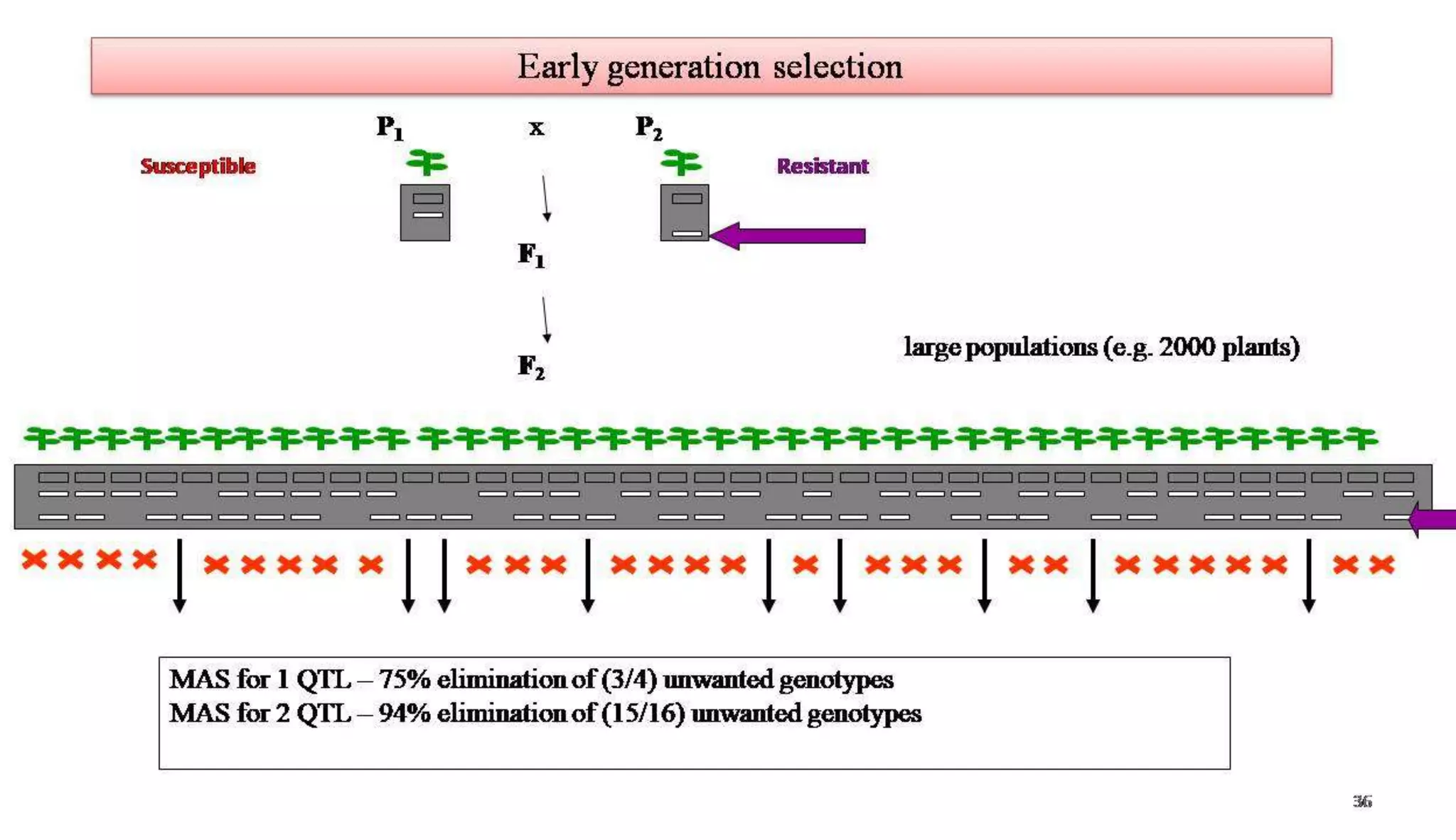
The document discusses marker-assisted selection (MAS) in plant breeding, highlighting its role in efficiently selecting desirable traits using molecular markers linked to specific genes. It outlines various types of markers, prerequisites for successful MAS programs, and its advantages such as reducing the time and cost of breeding. Additionally, it addresses the limitations of MAS and details different breeding schemes, including marker-assisted backcrossing, gene pyramiding, and early generation selection.