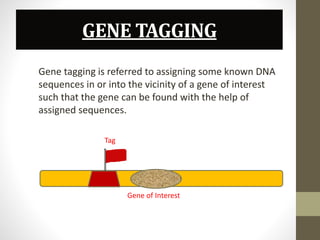
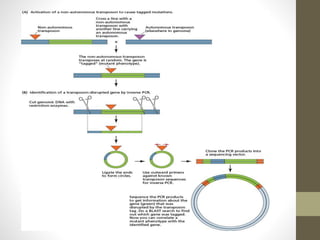
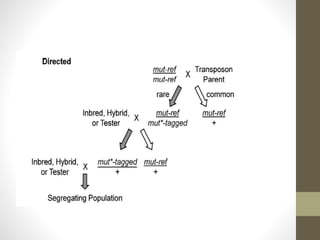
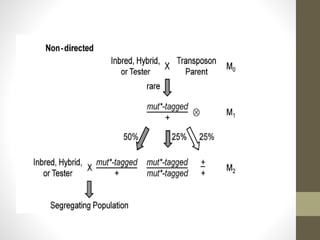
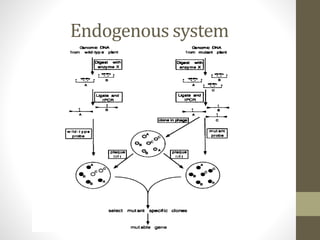
This document summarizes transposon tagging as a method to identify genes. Transposon tagging involves inserting a transposon near a gene of interest, which then allows the gene to be identified based on its proximity to the transposon. The document discusses different types of transposons used for tagging in plants and animals. It describes approaches for both targeted and non-targeted tagging and methods for identifying the tagged gene, including RFLP analysis and inverse PCR. As an example, it summarizes how the Cf-9 gene conferring resistance to leaf mold in tomato was identified using Ds transposon tagging.