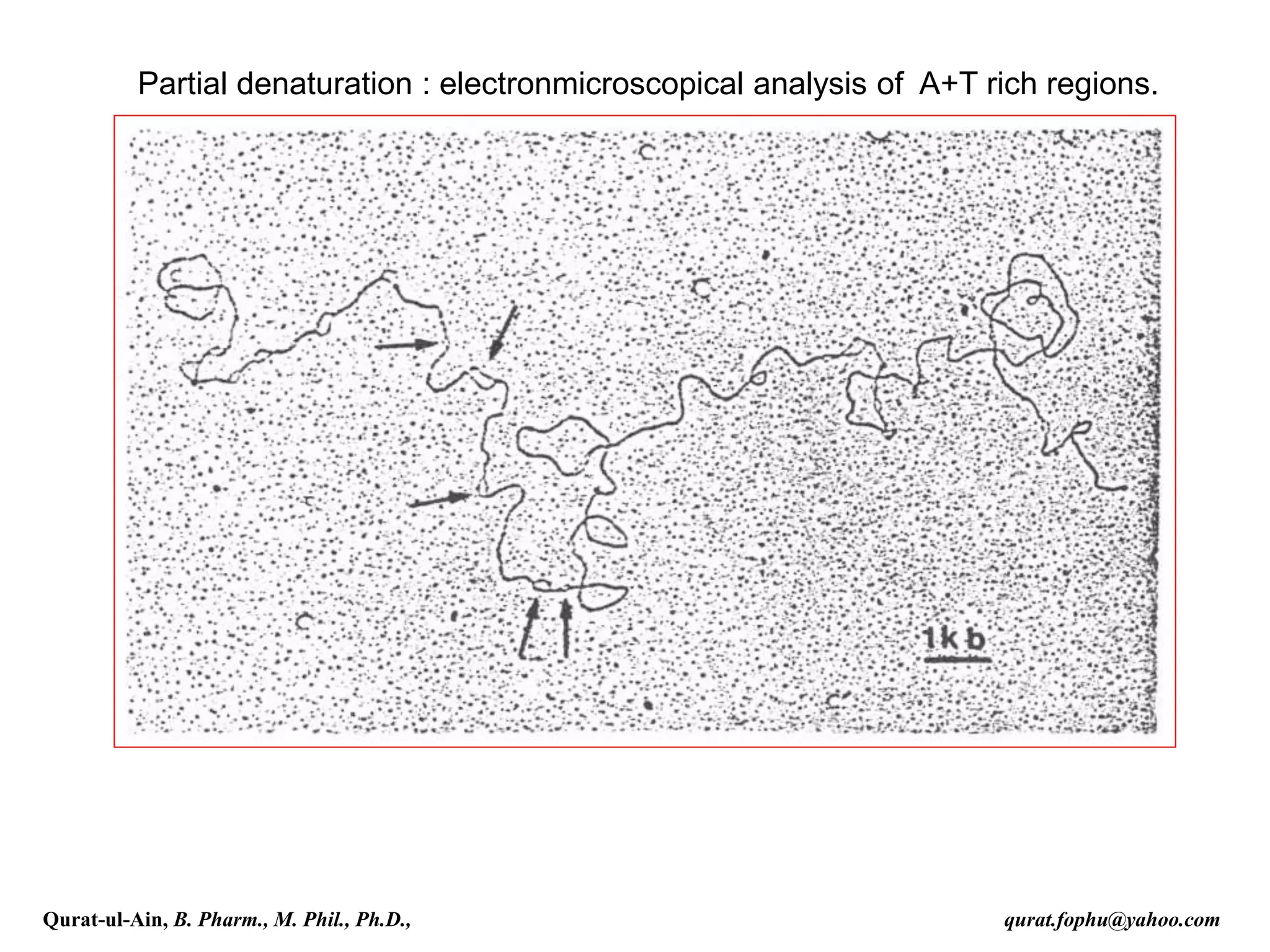
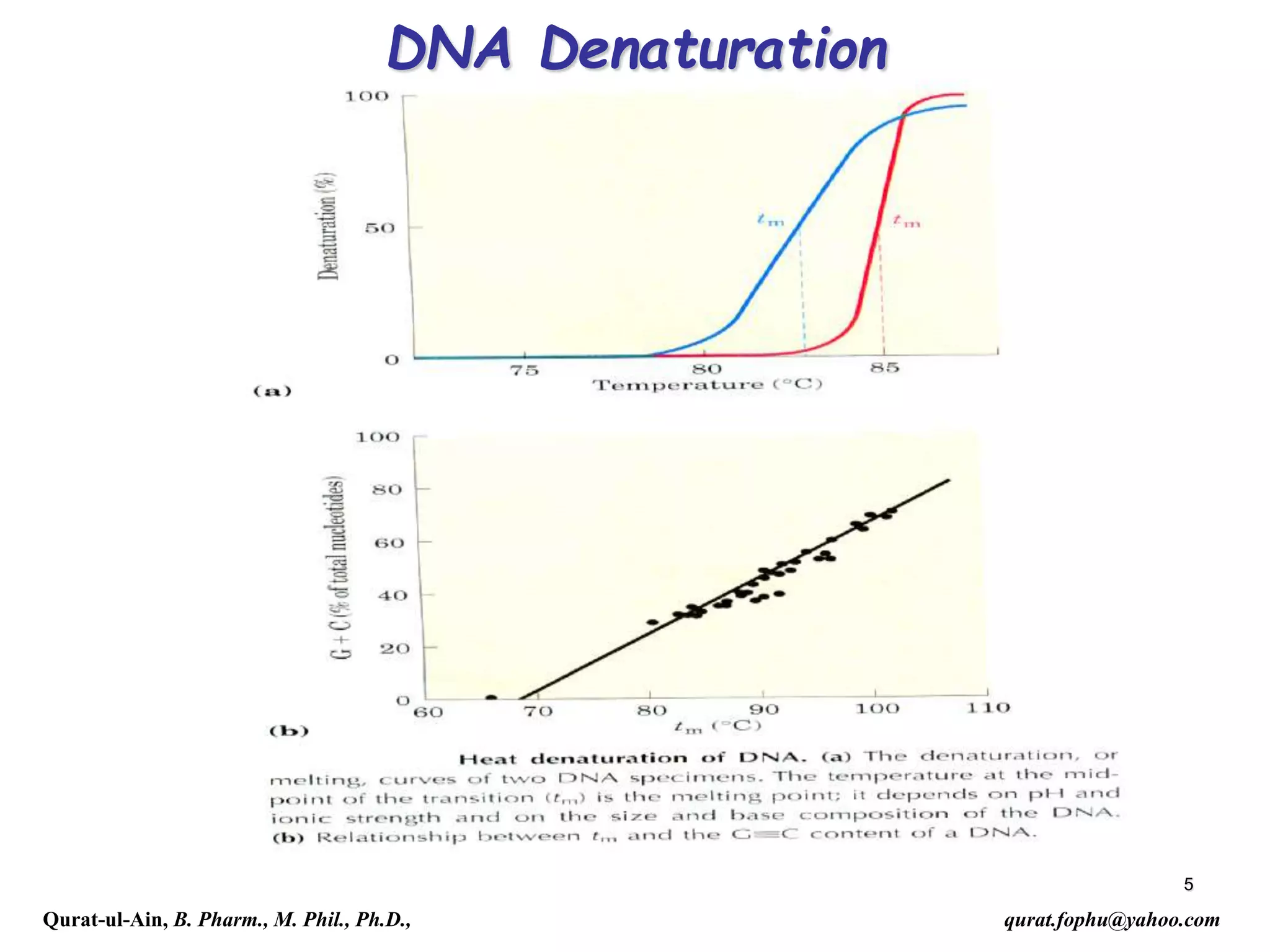
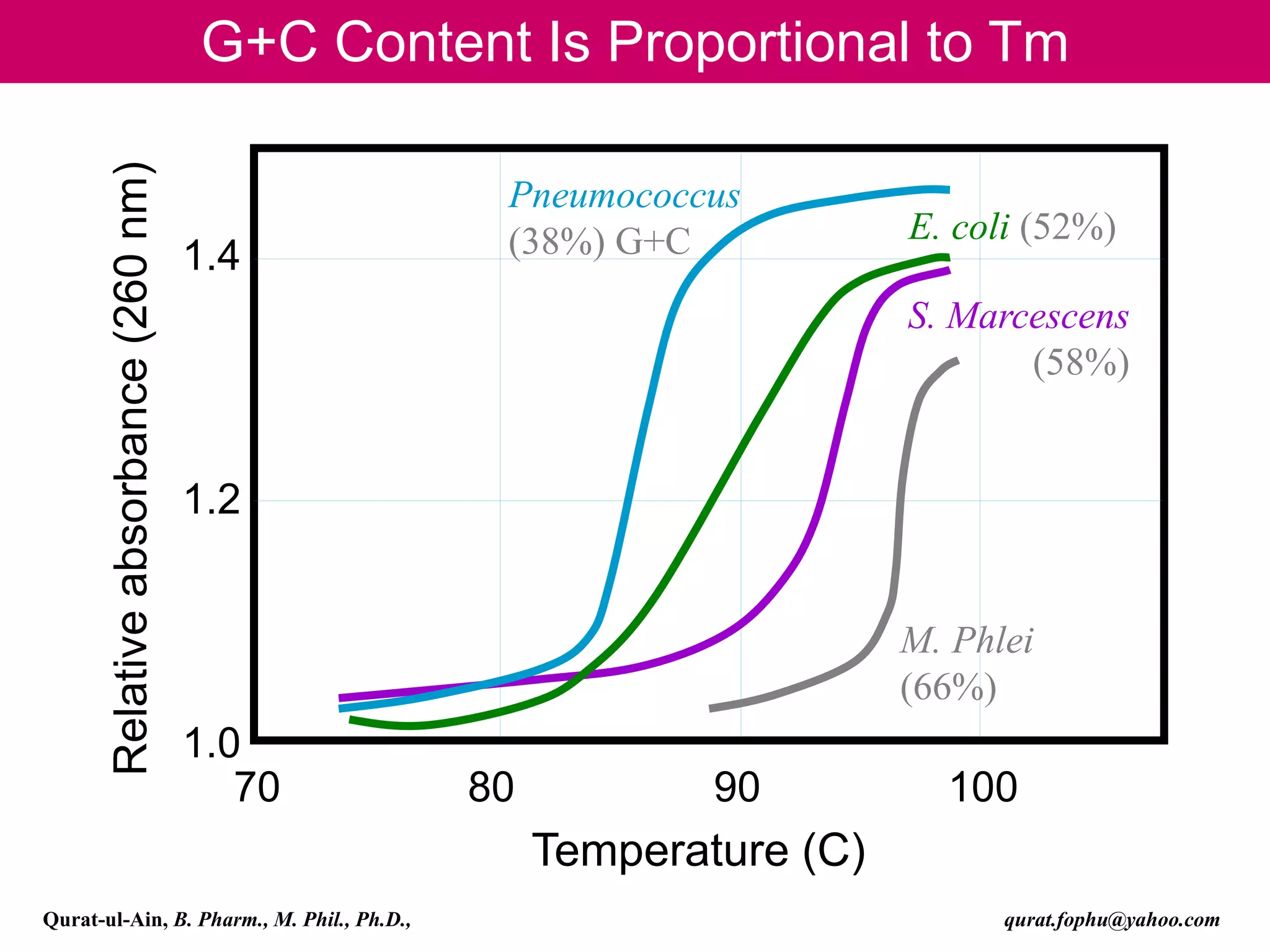
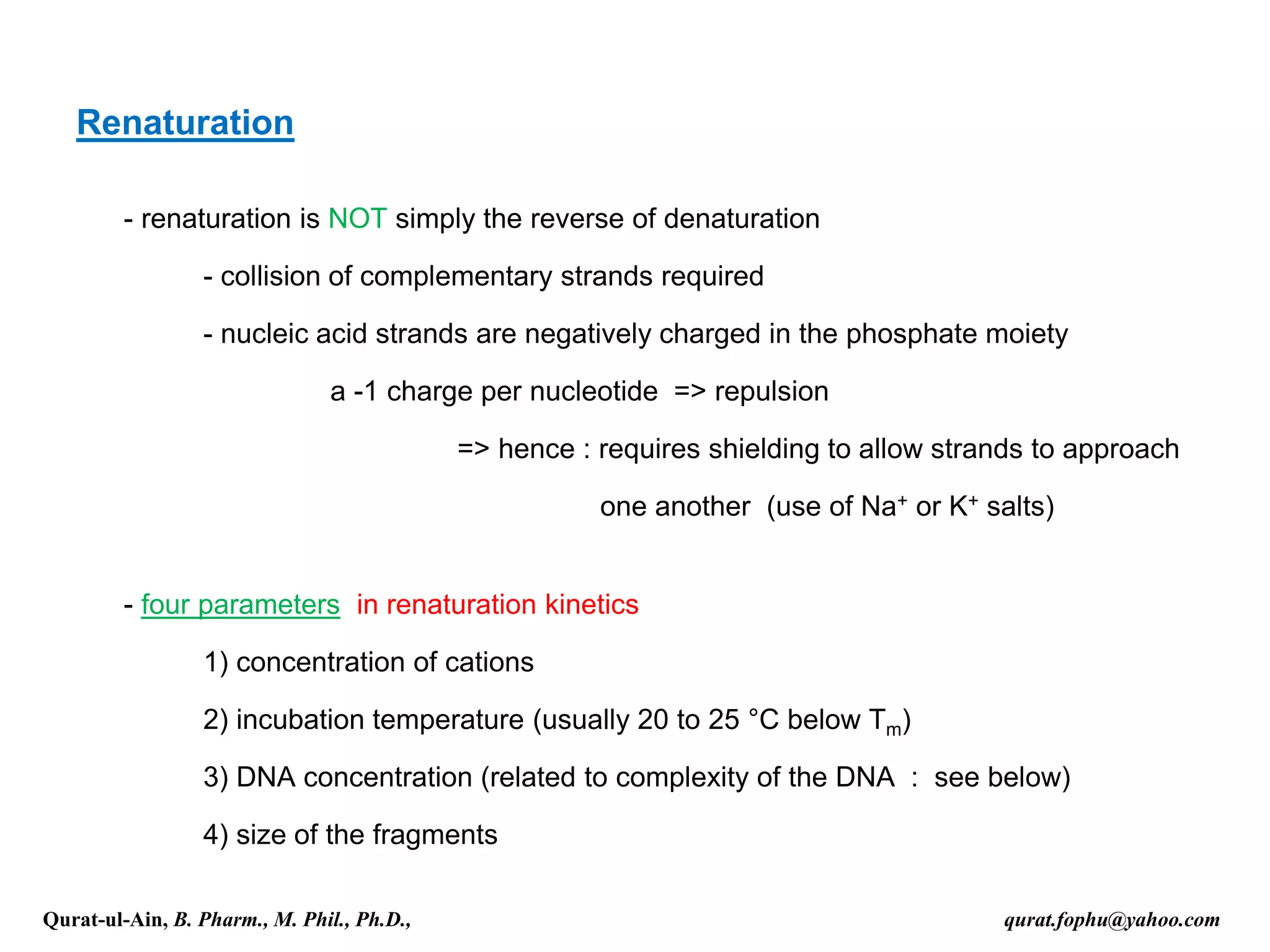
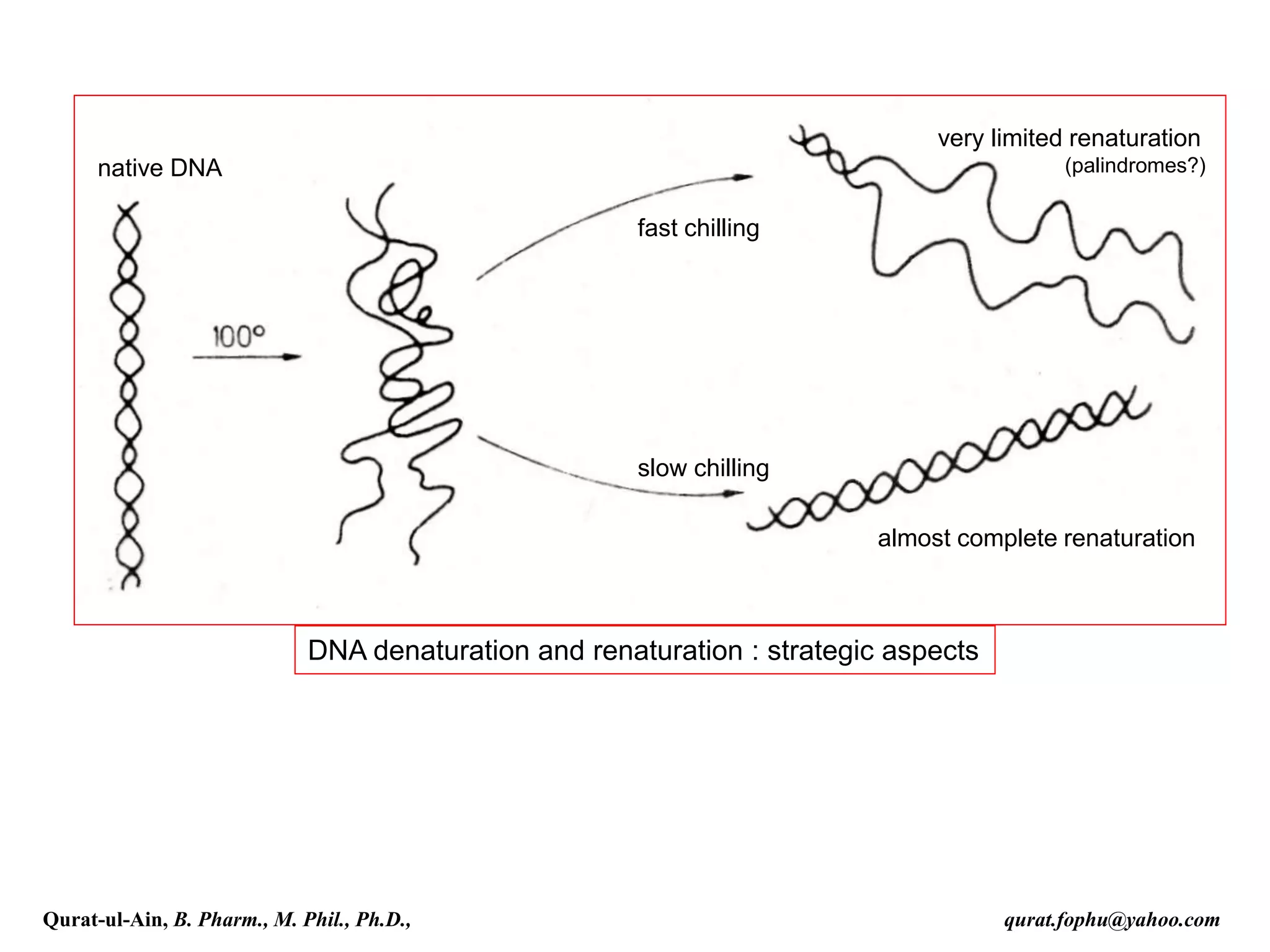
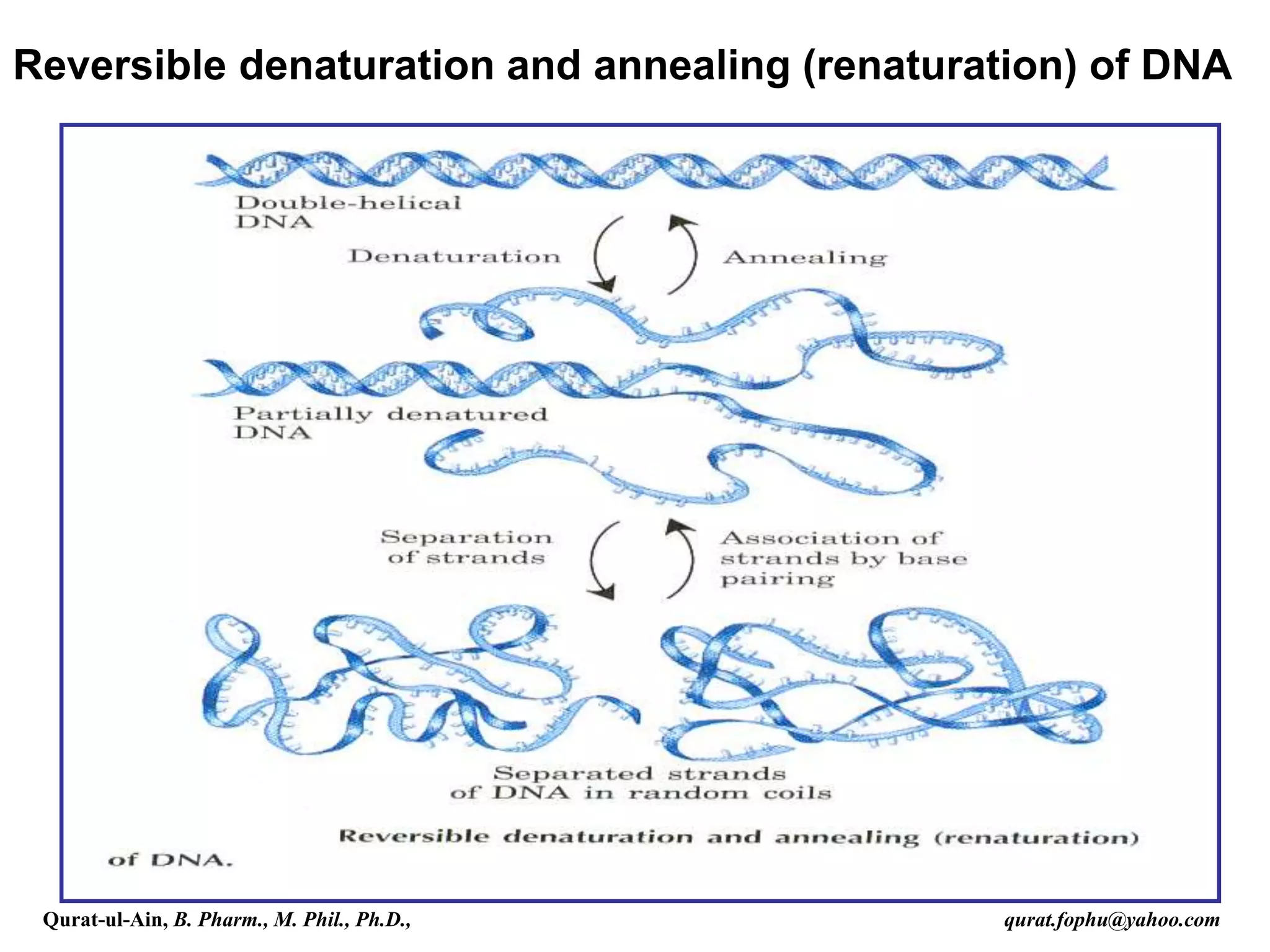
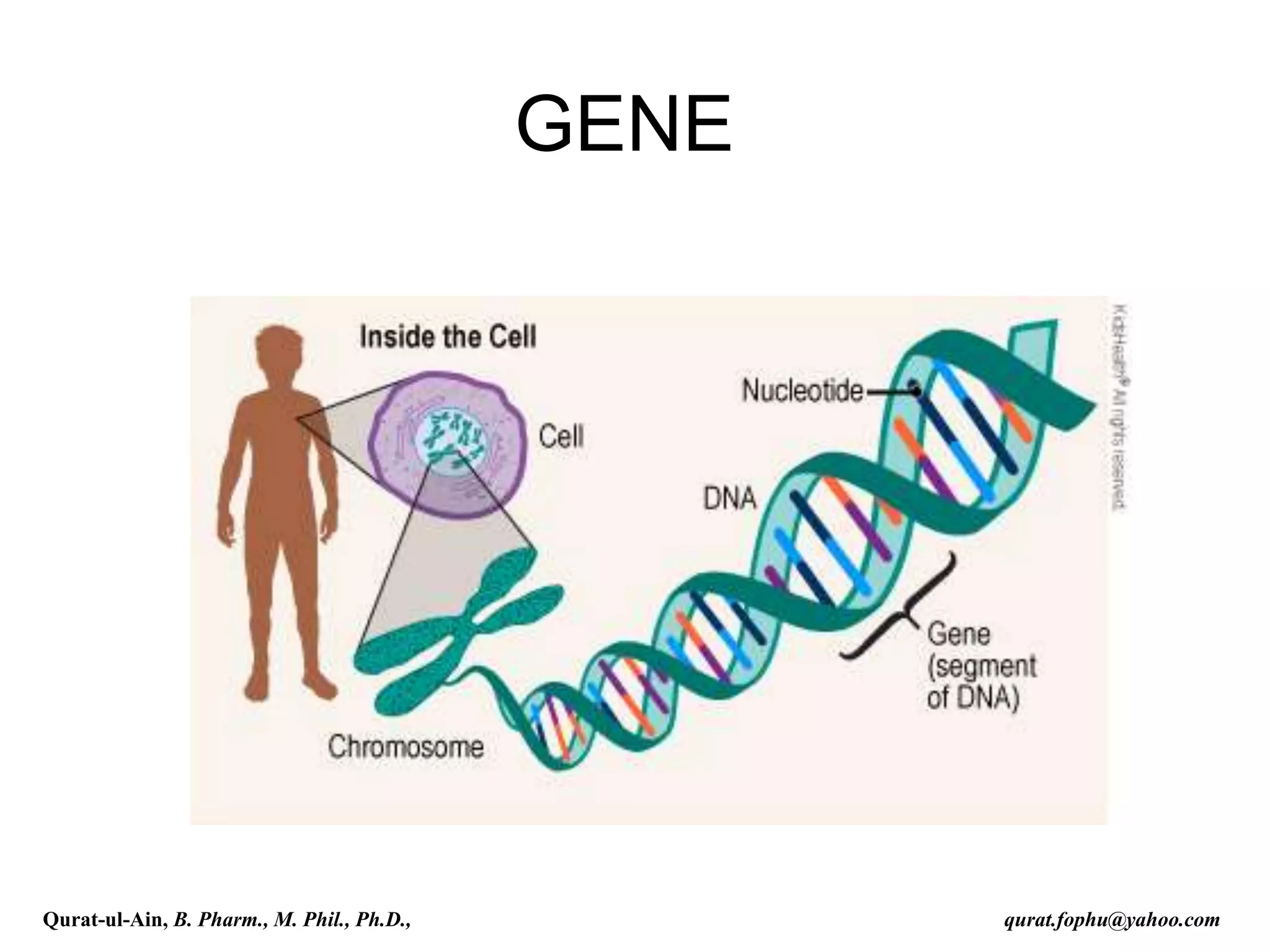
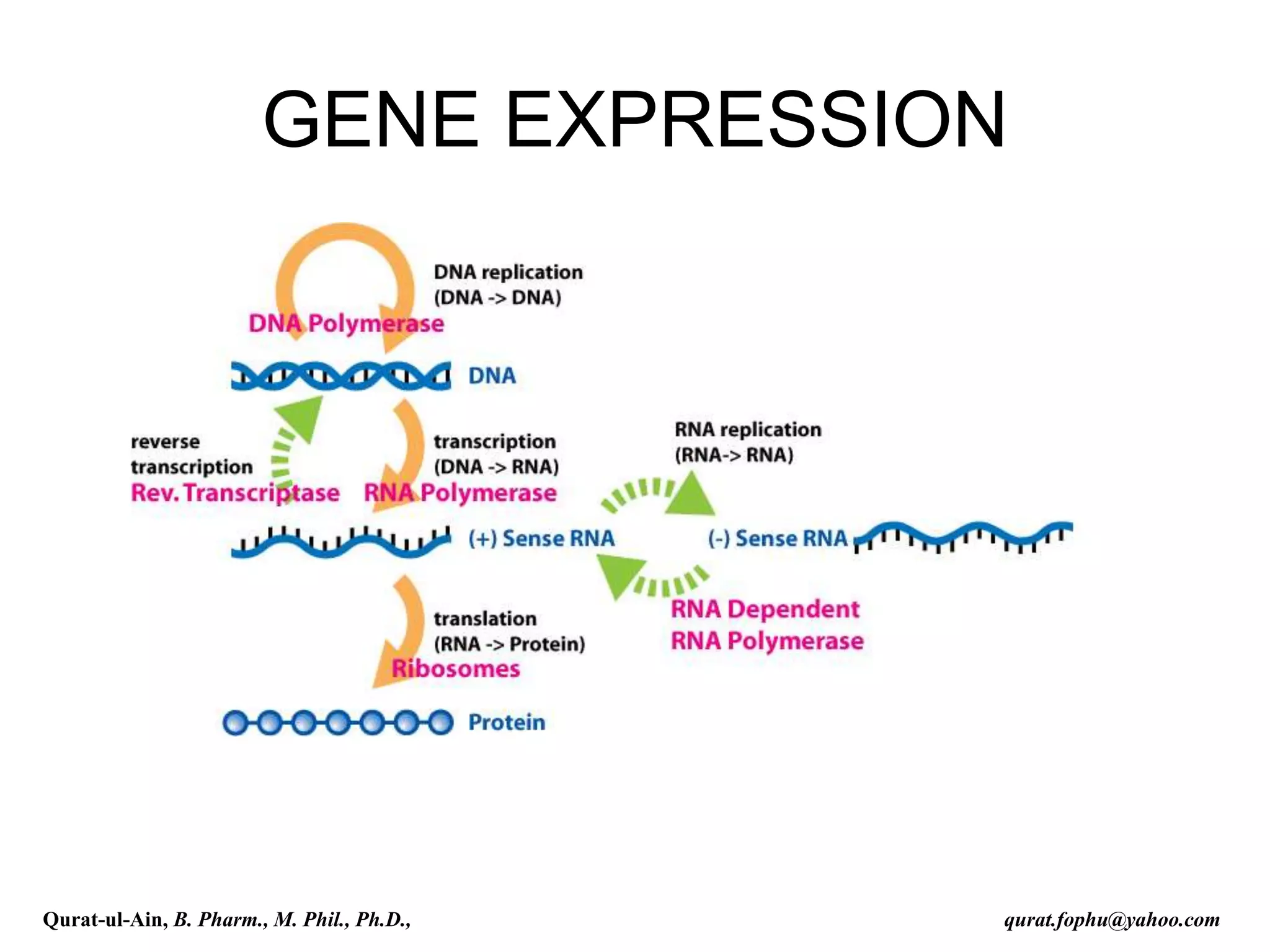
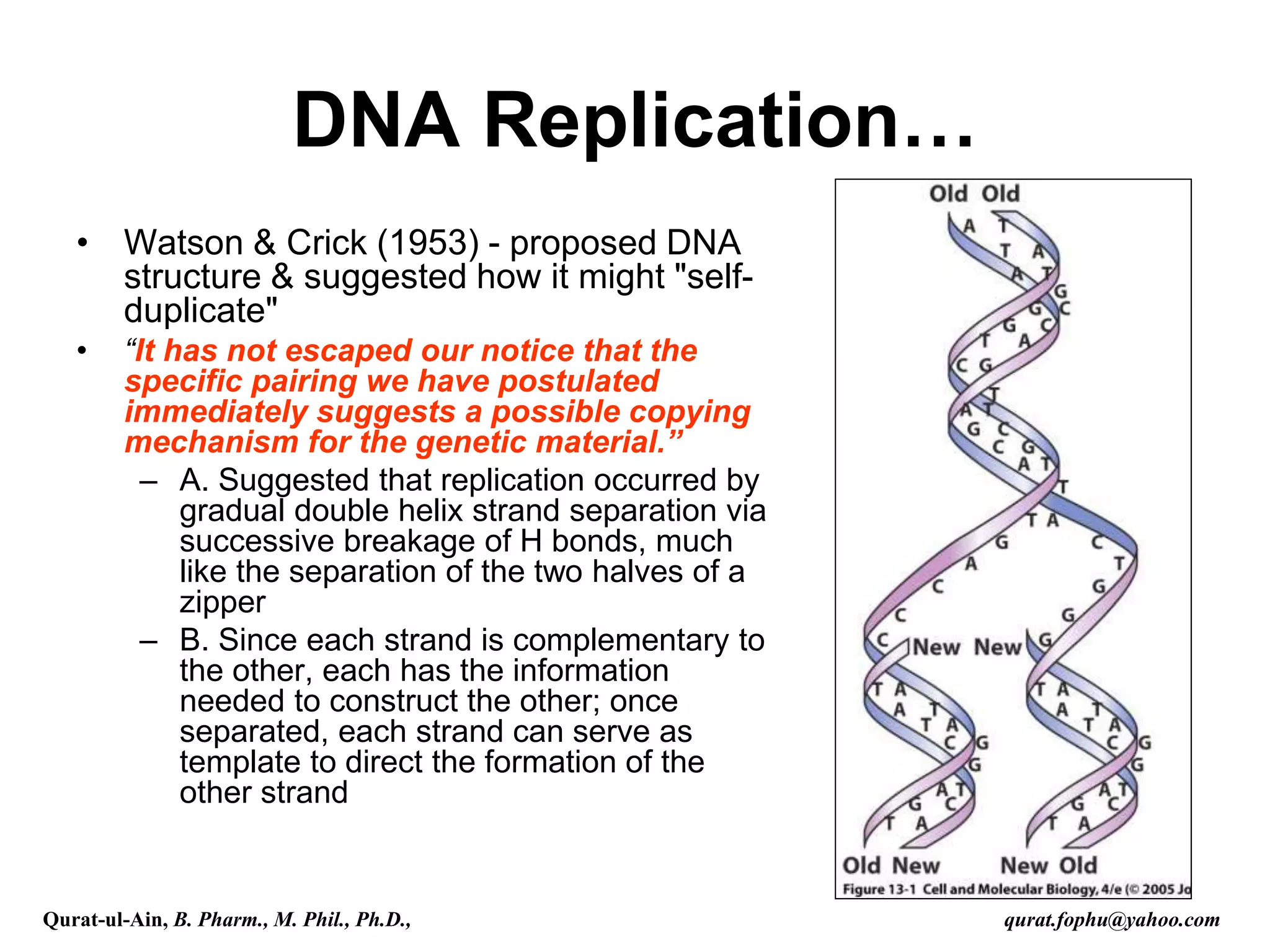
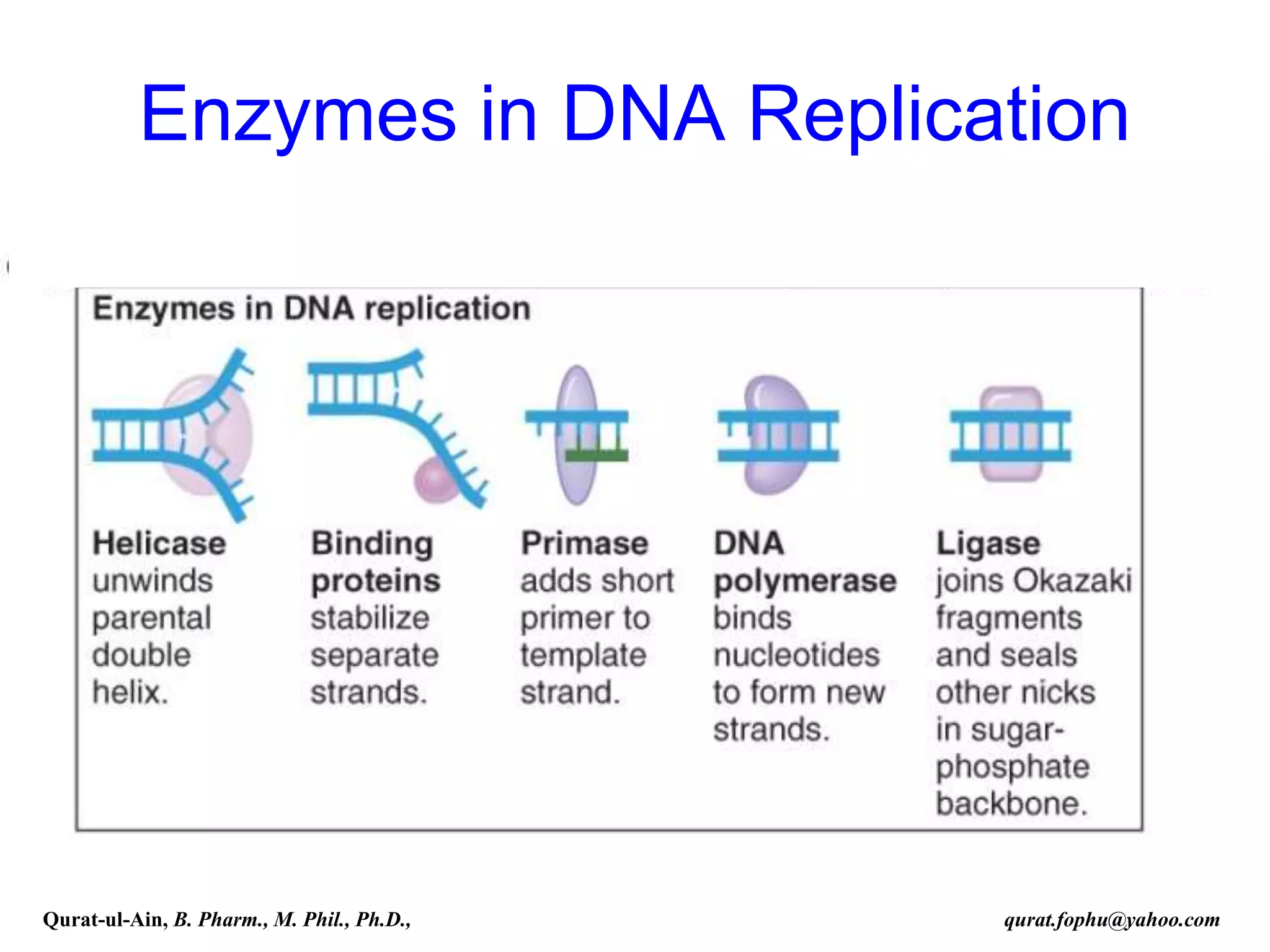
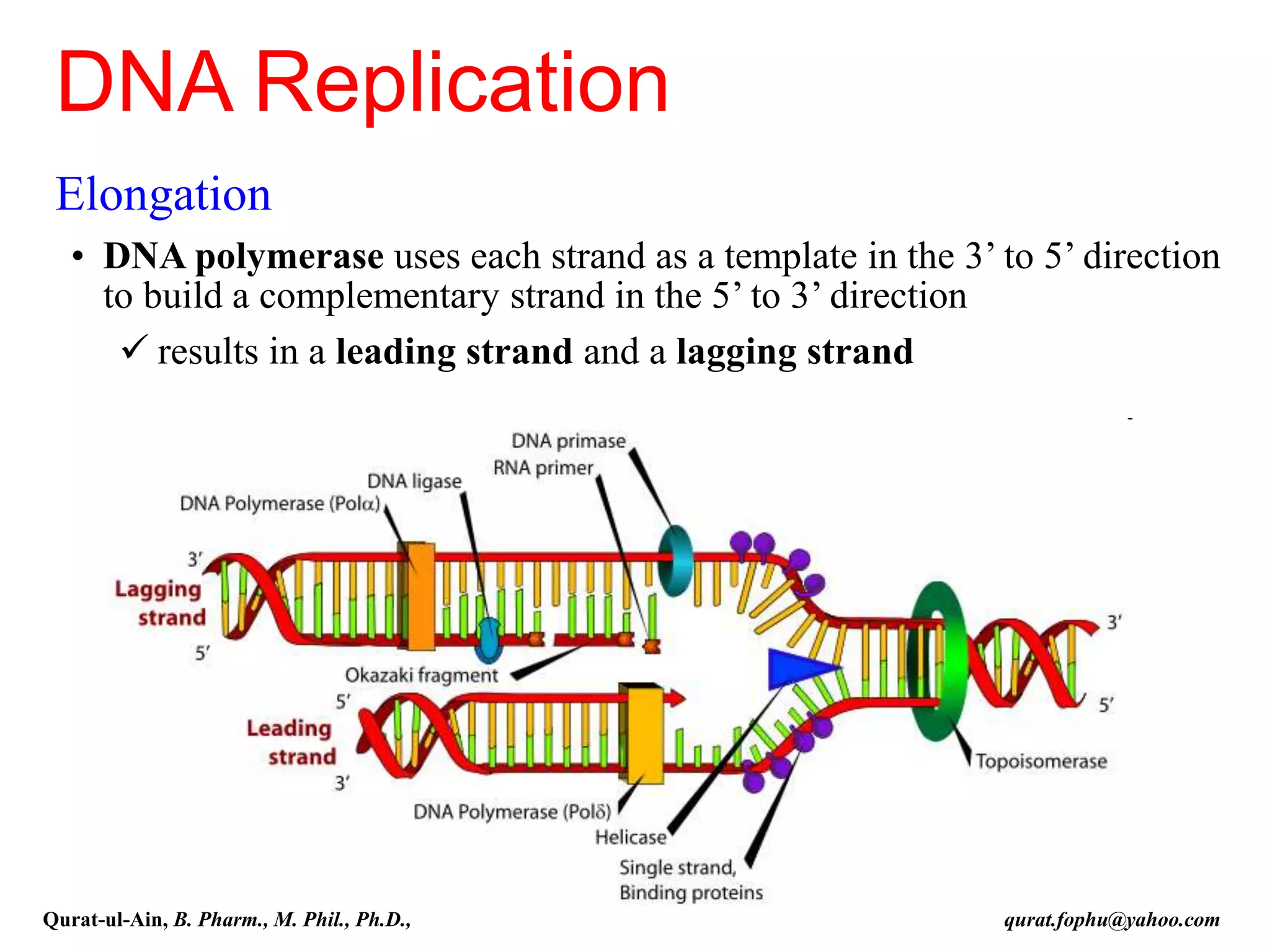
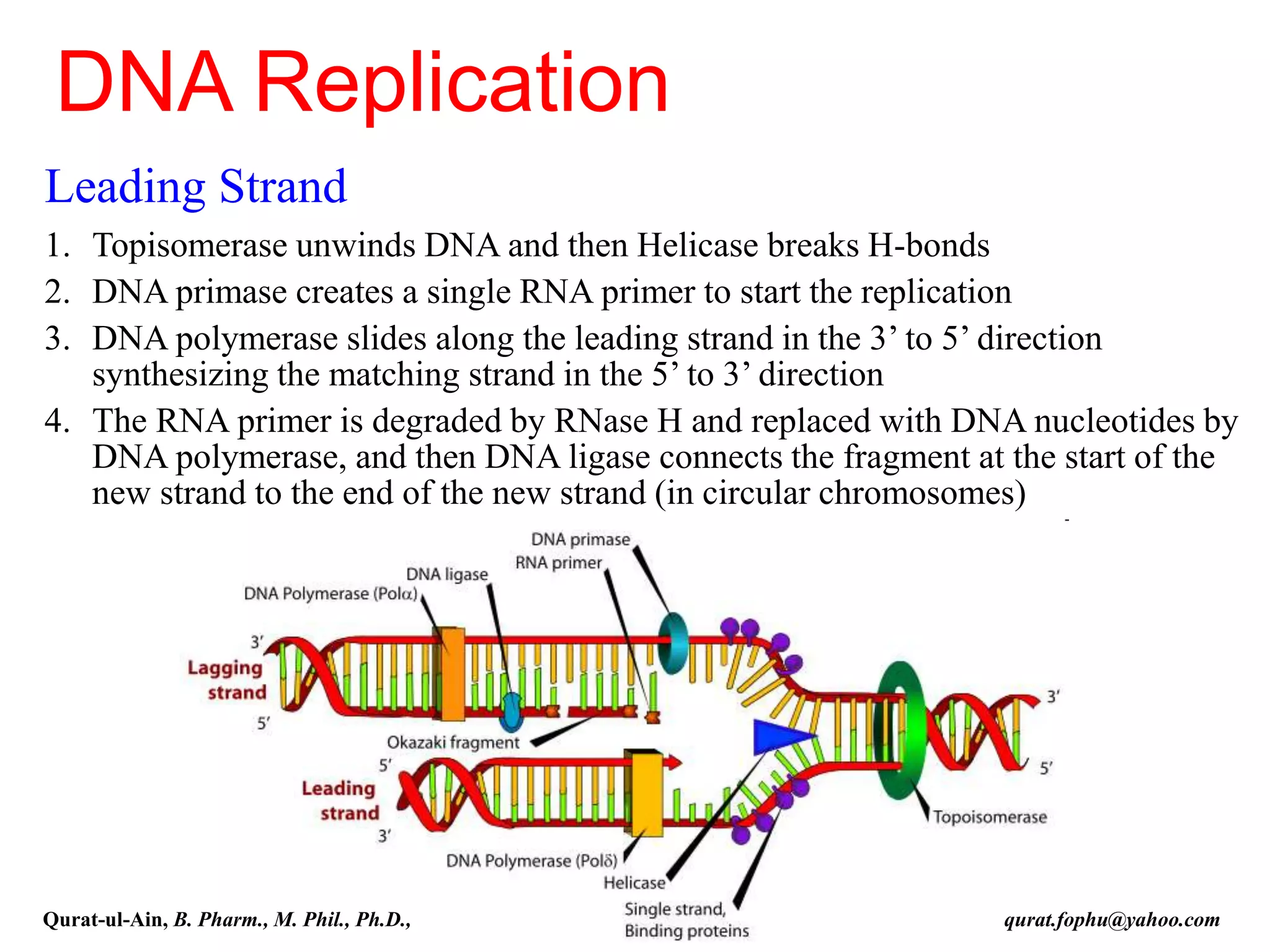
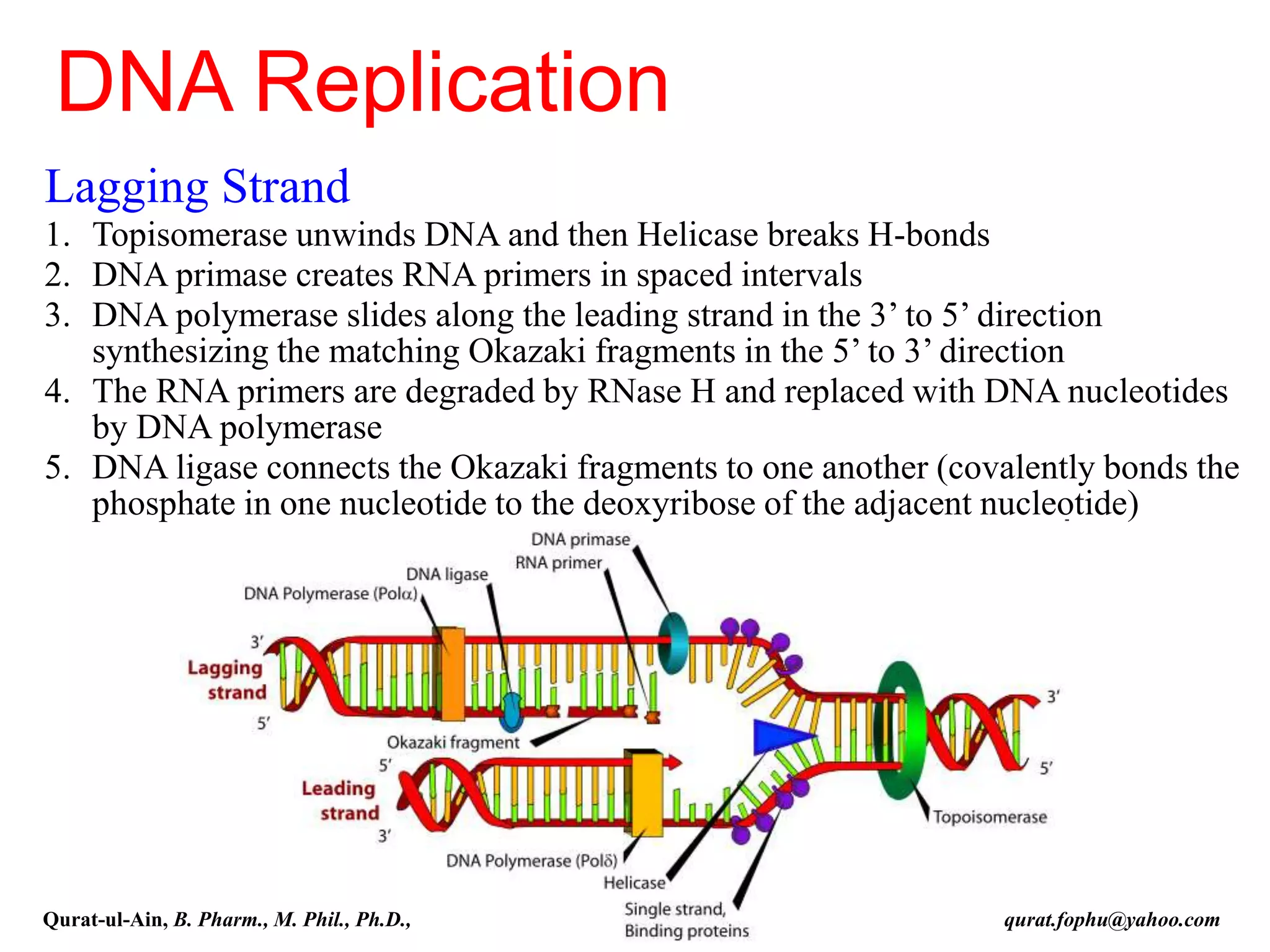
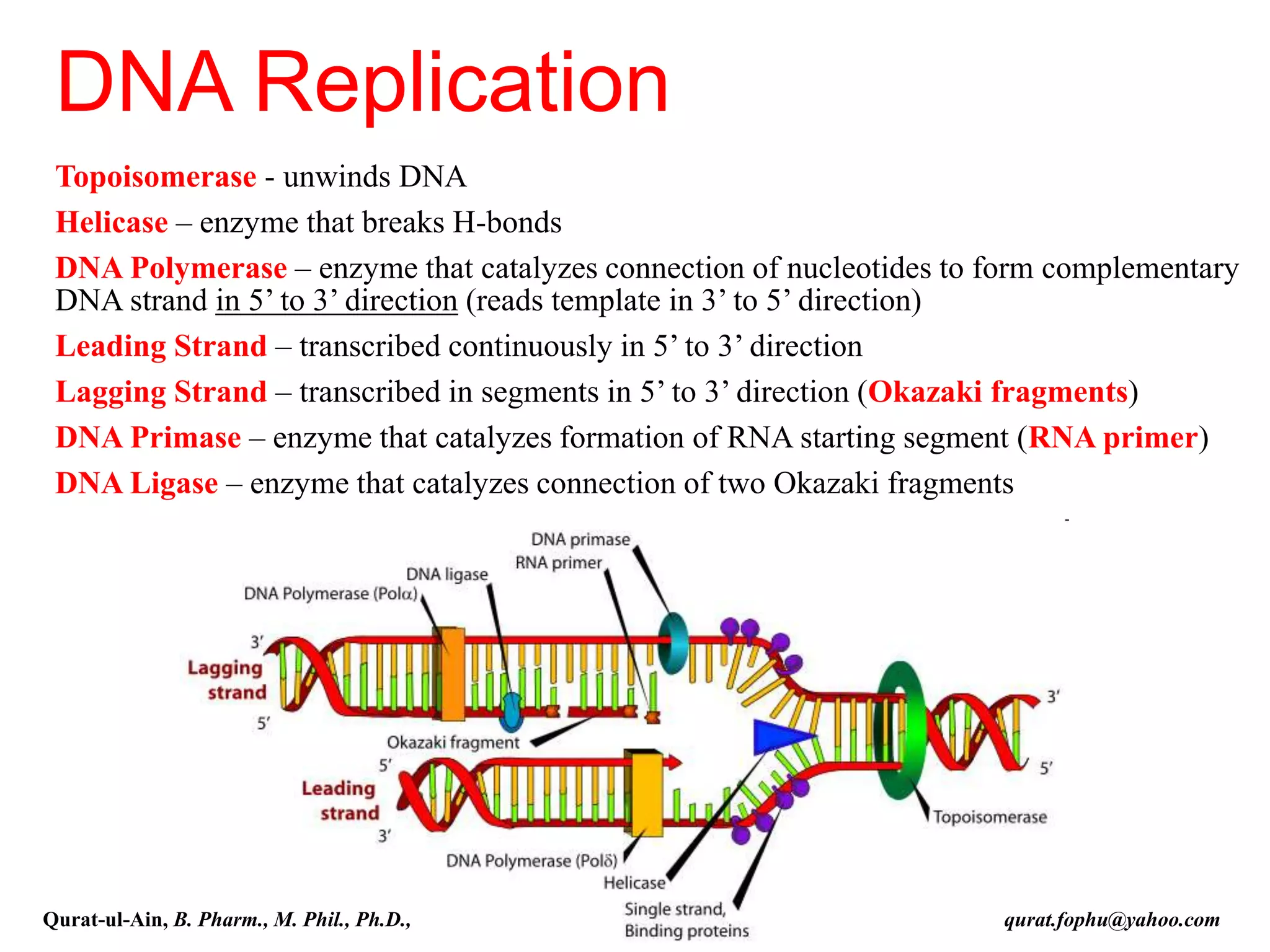
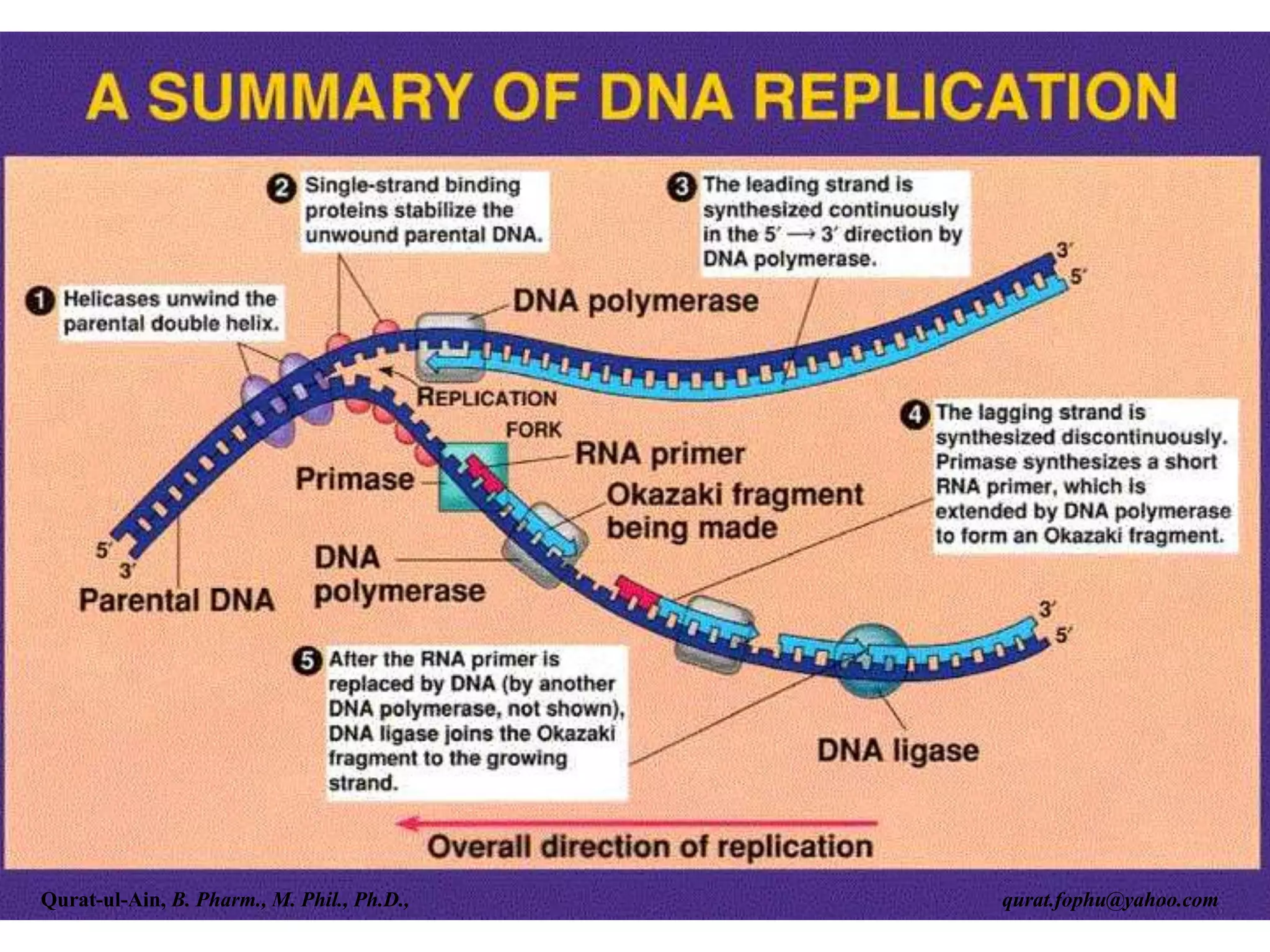
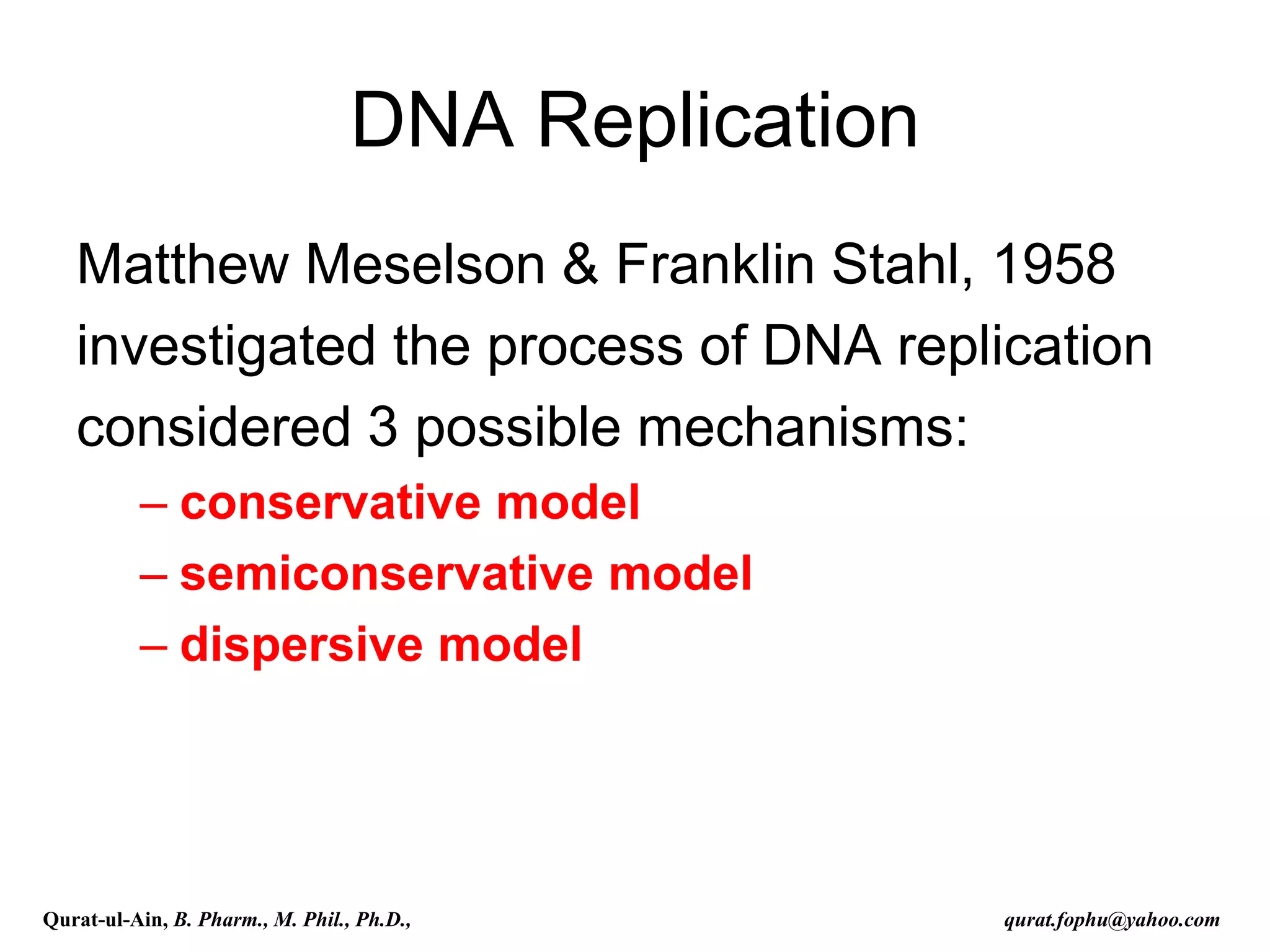
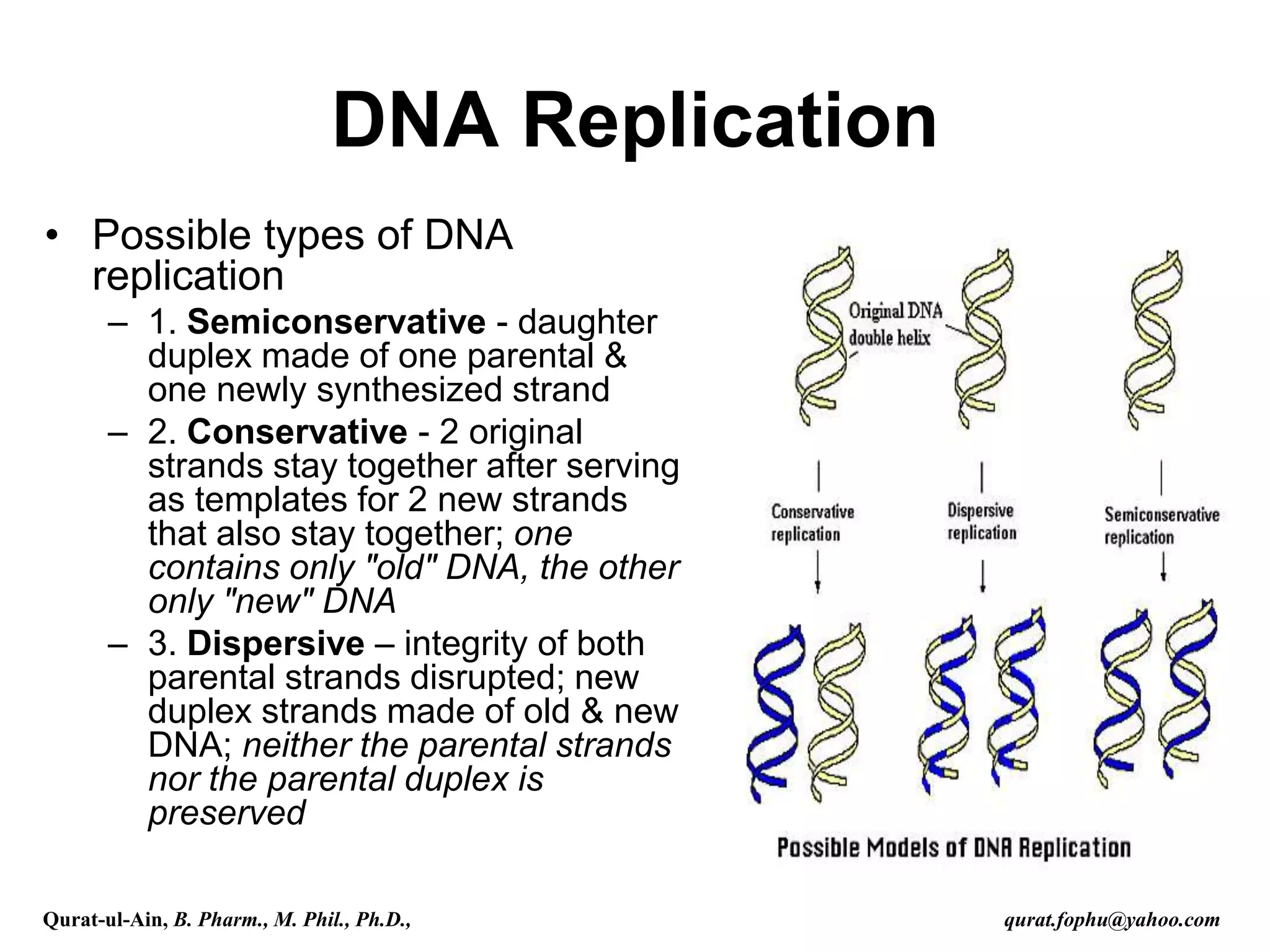
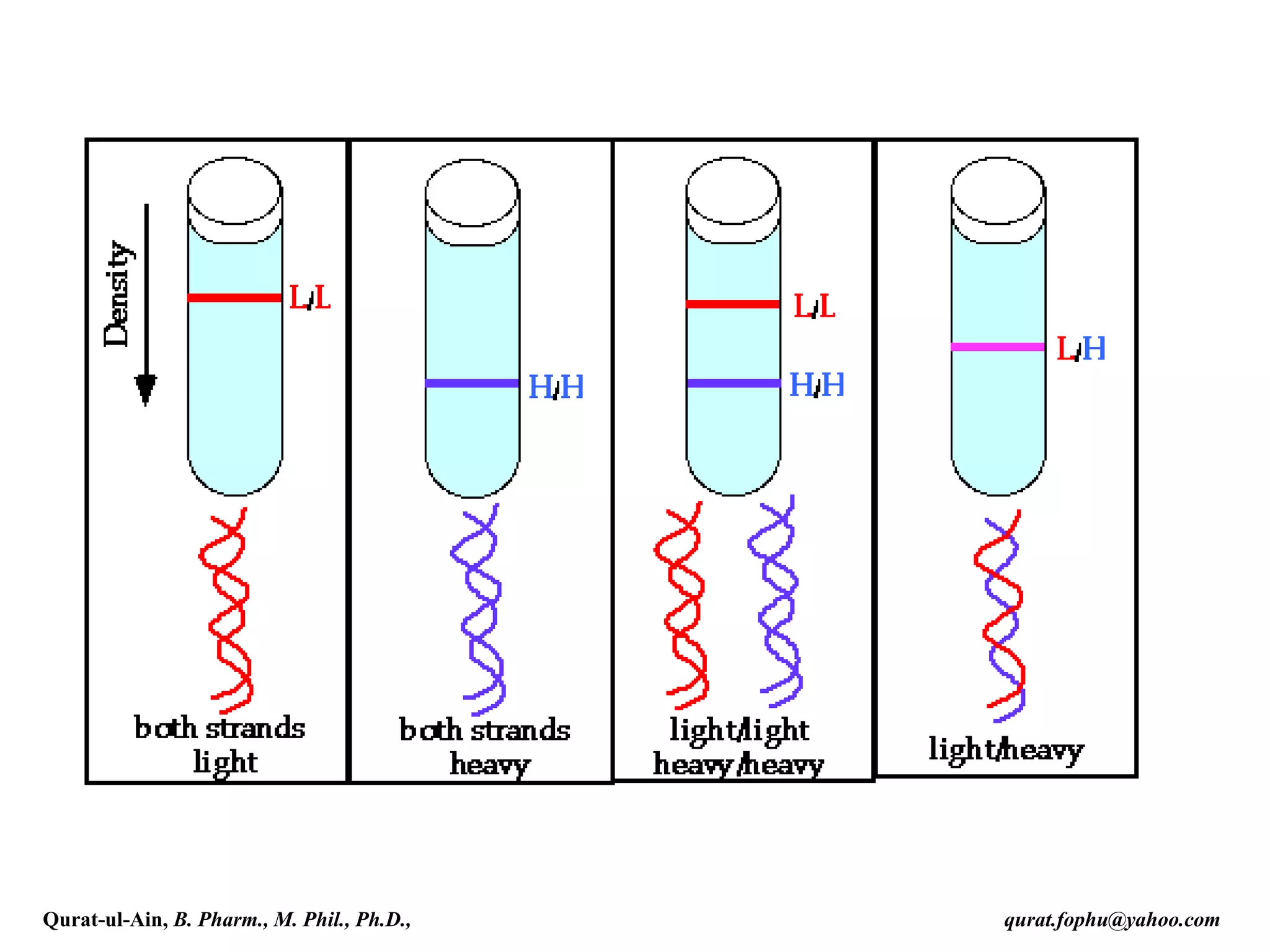
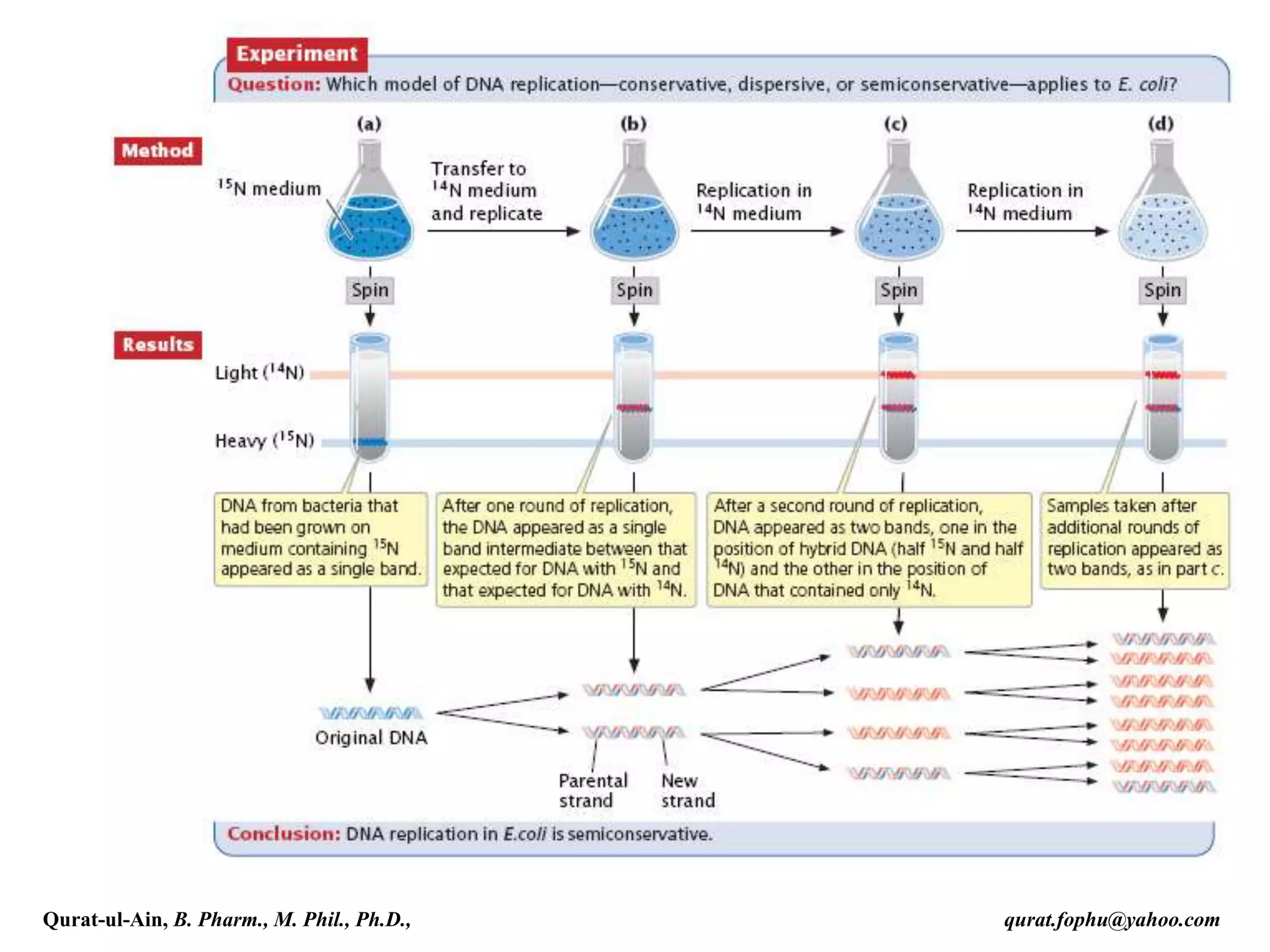
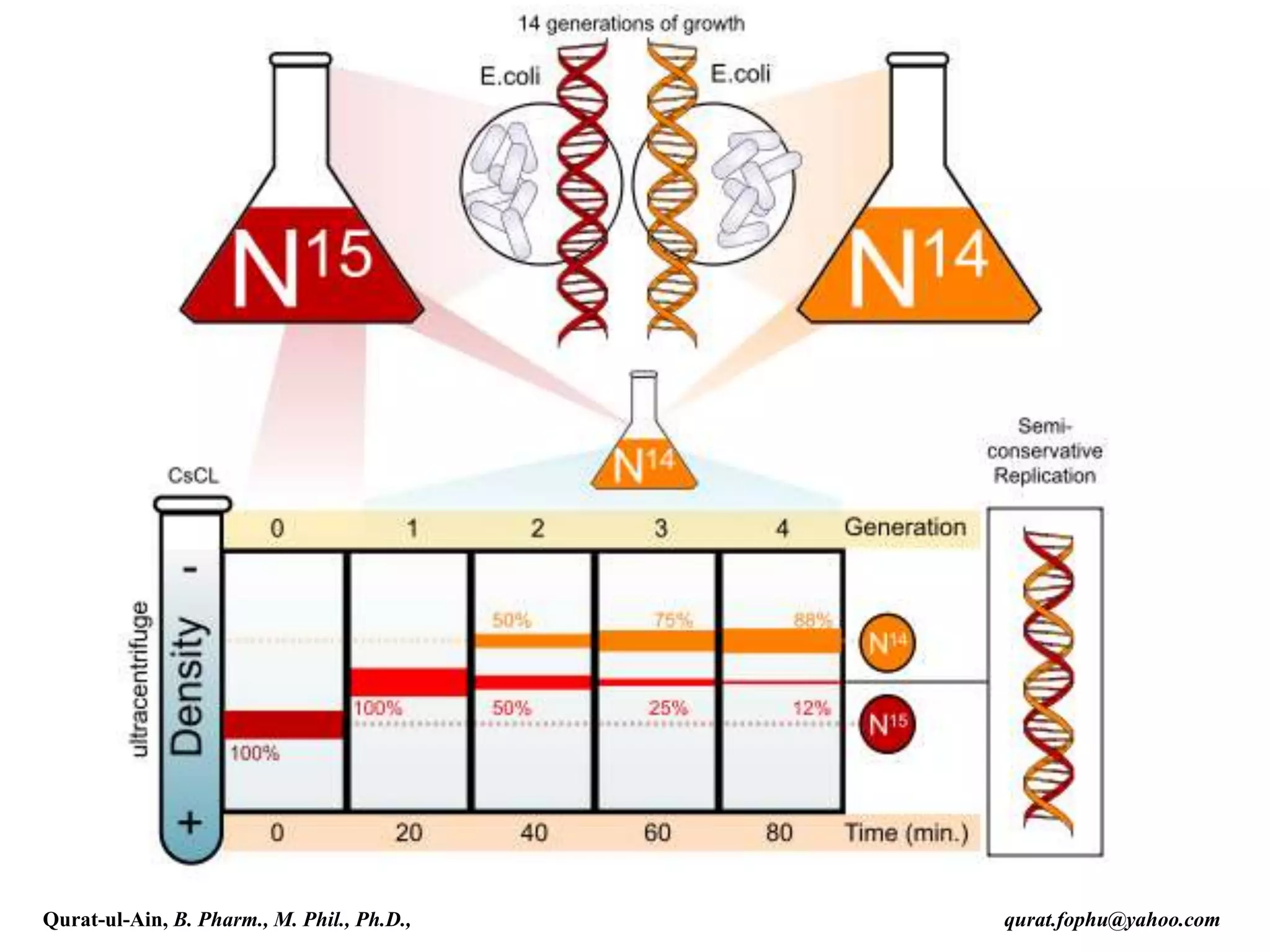
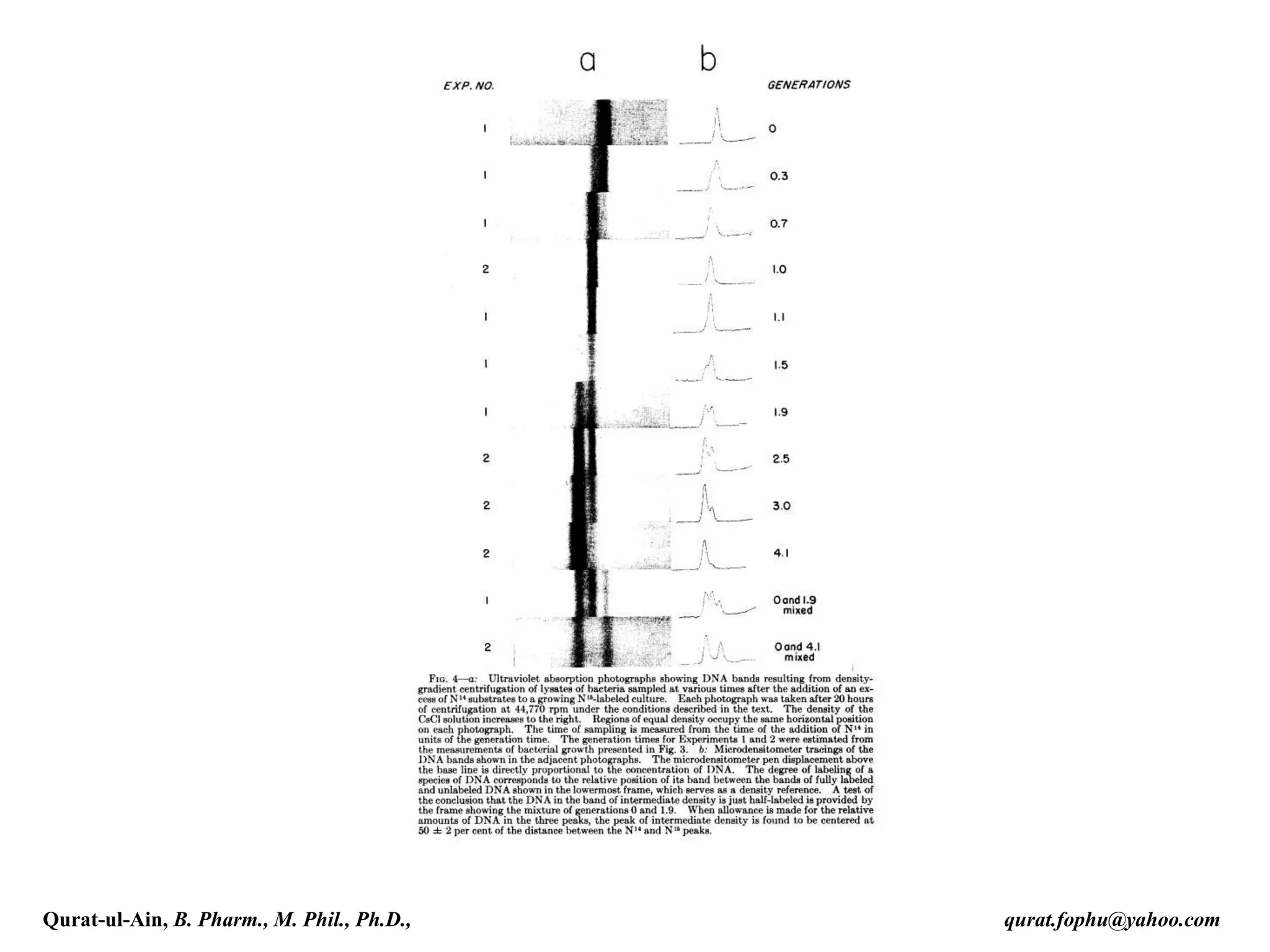
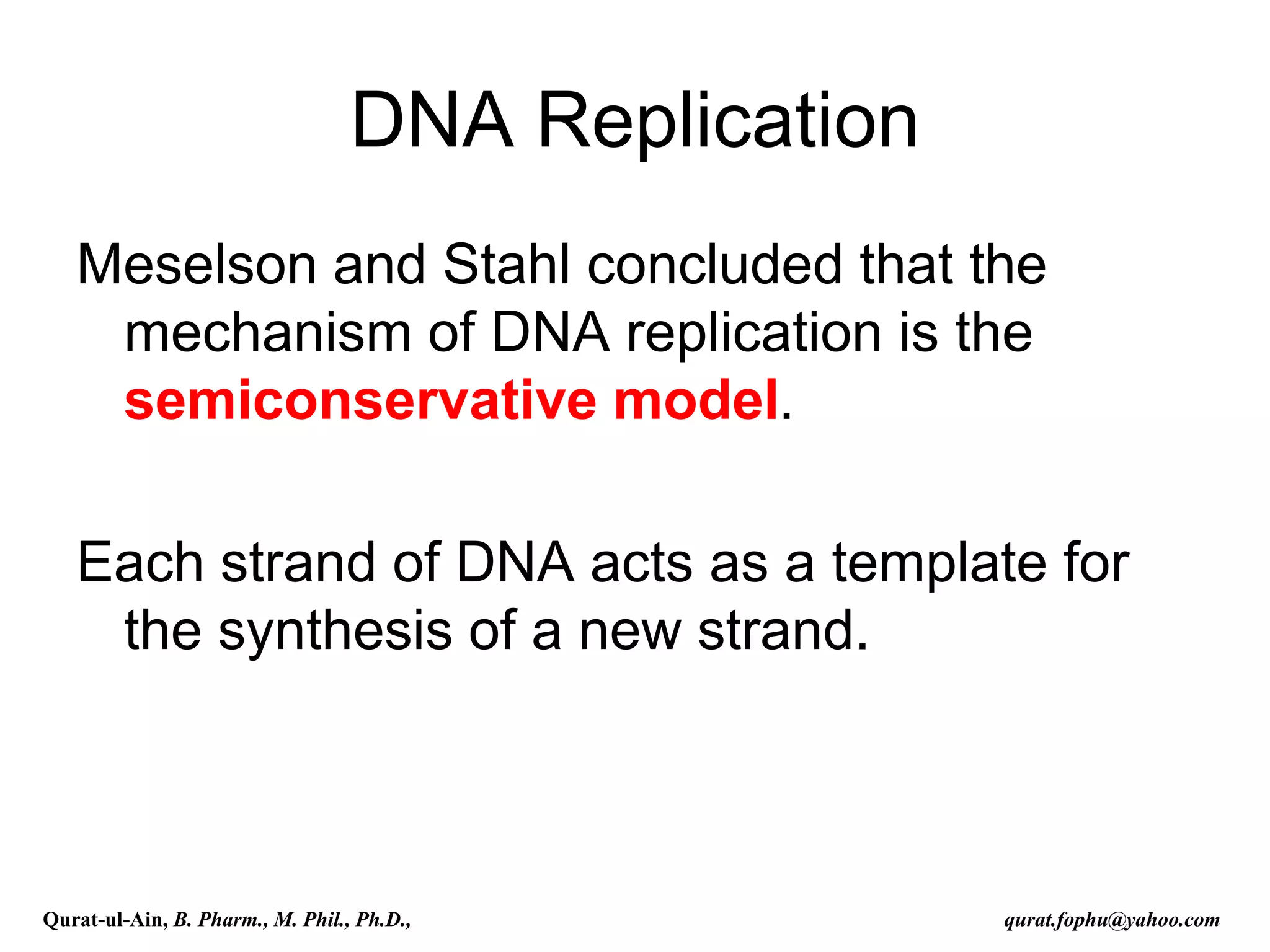
The document discusses denaturation and renaturation of DNA and replication of DNA. It provides details on how DNA can be denatured using heat, chemicals, or proteins. Renaturation requires energy and slow cooling to allow base pairs to rebuild. DNA replication is semiconservative, with each strand serving as a template to produce a new complementary strand. Meselson and Stahl's experiment demonstrated that replication is semiconservative through detection of bands at different densities on centrifugation.