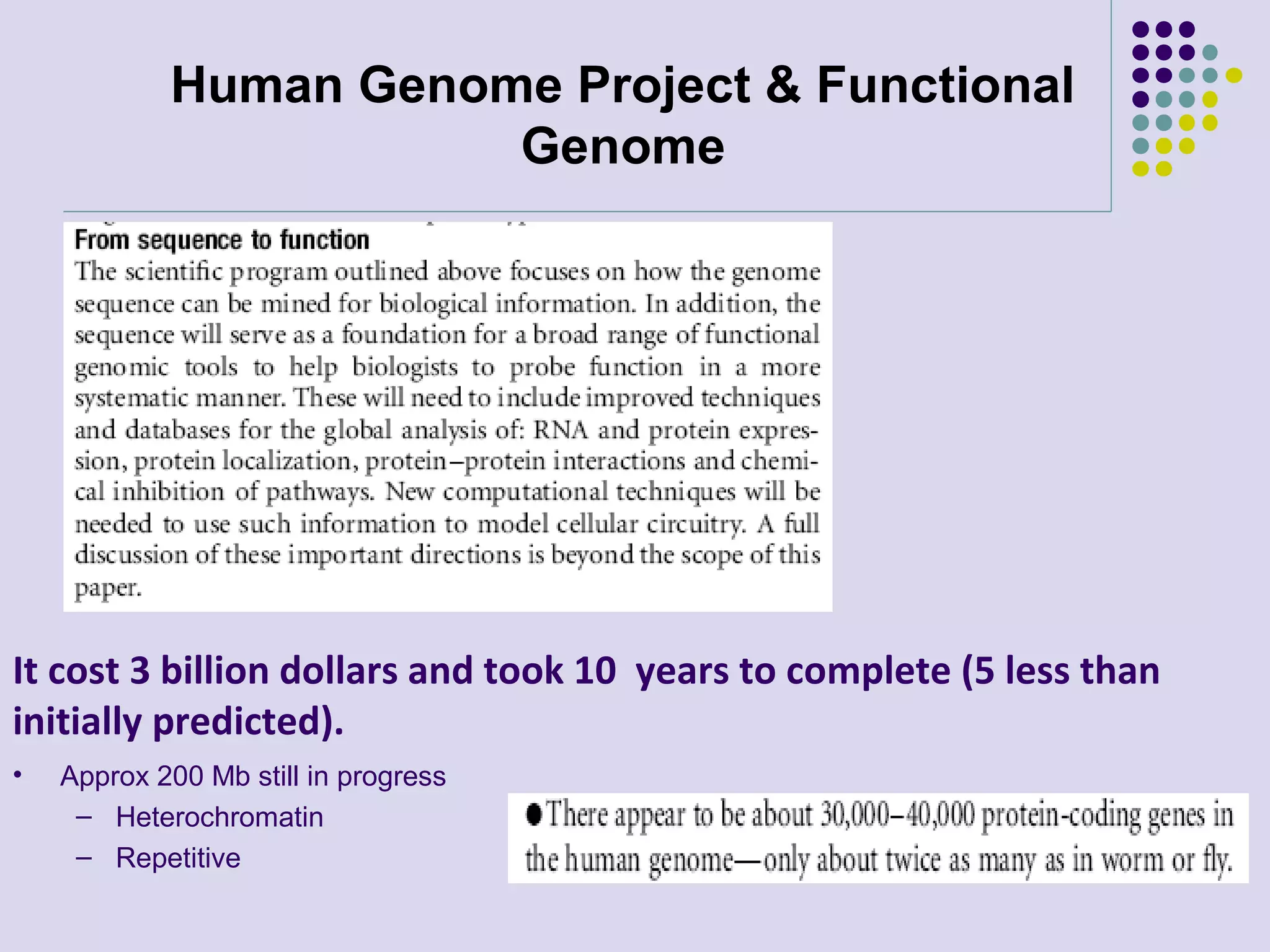
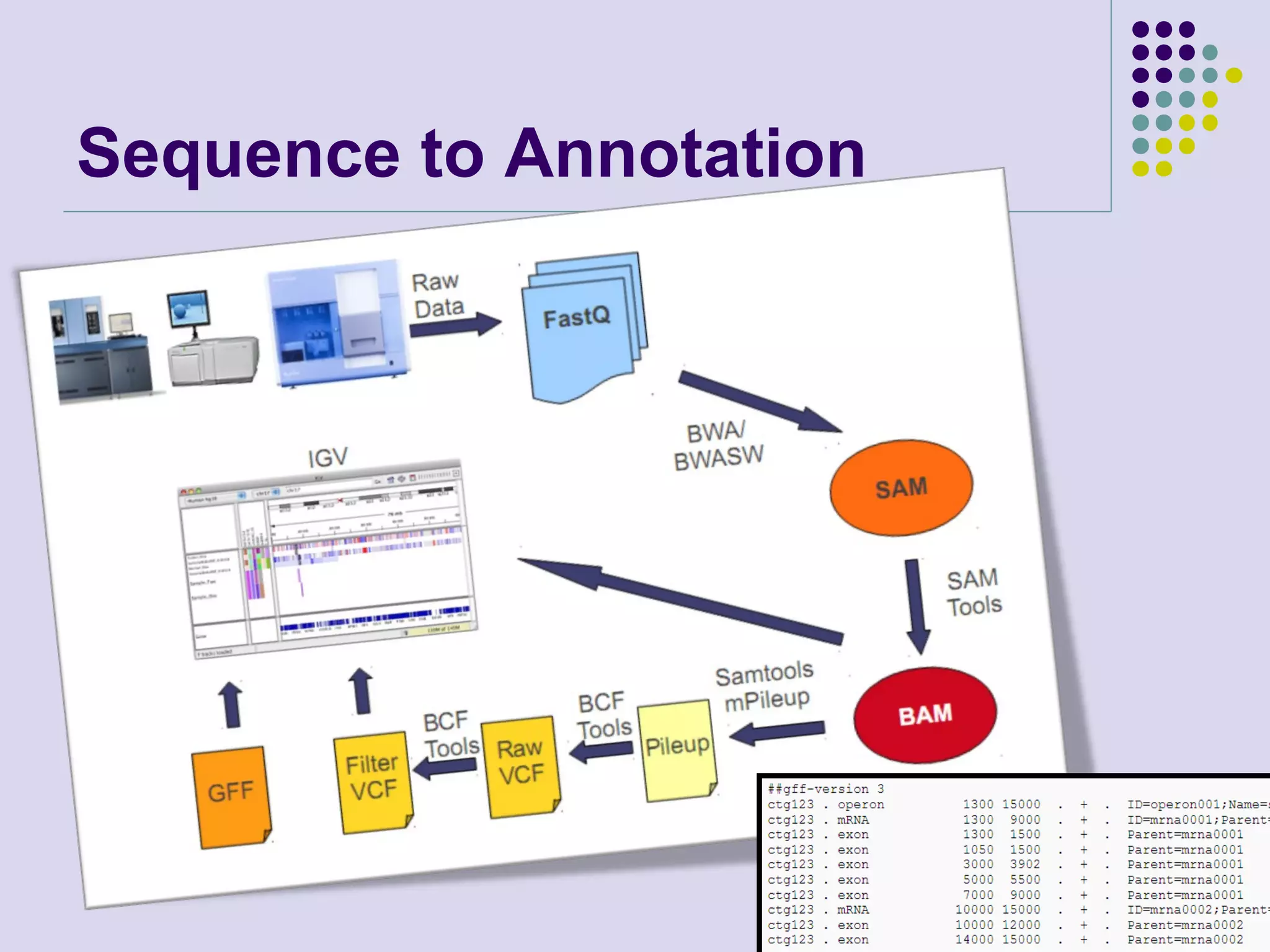
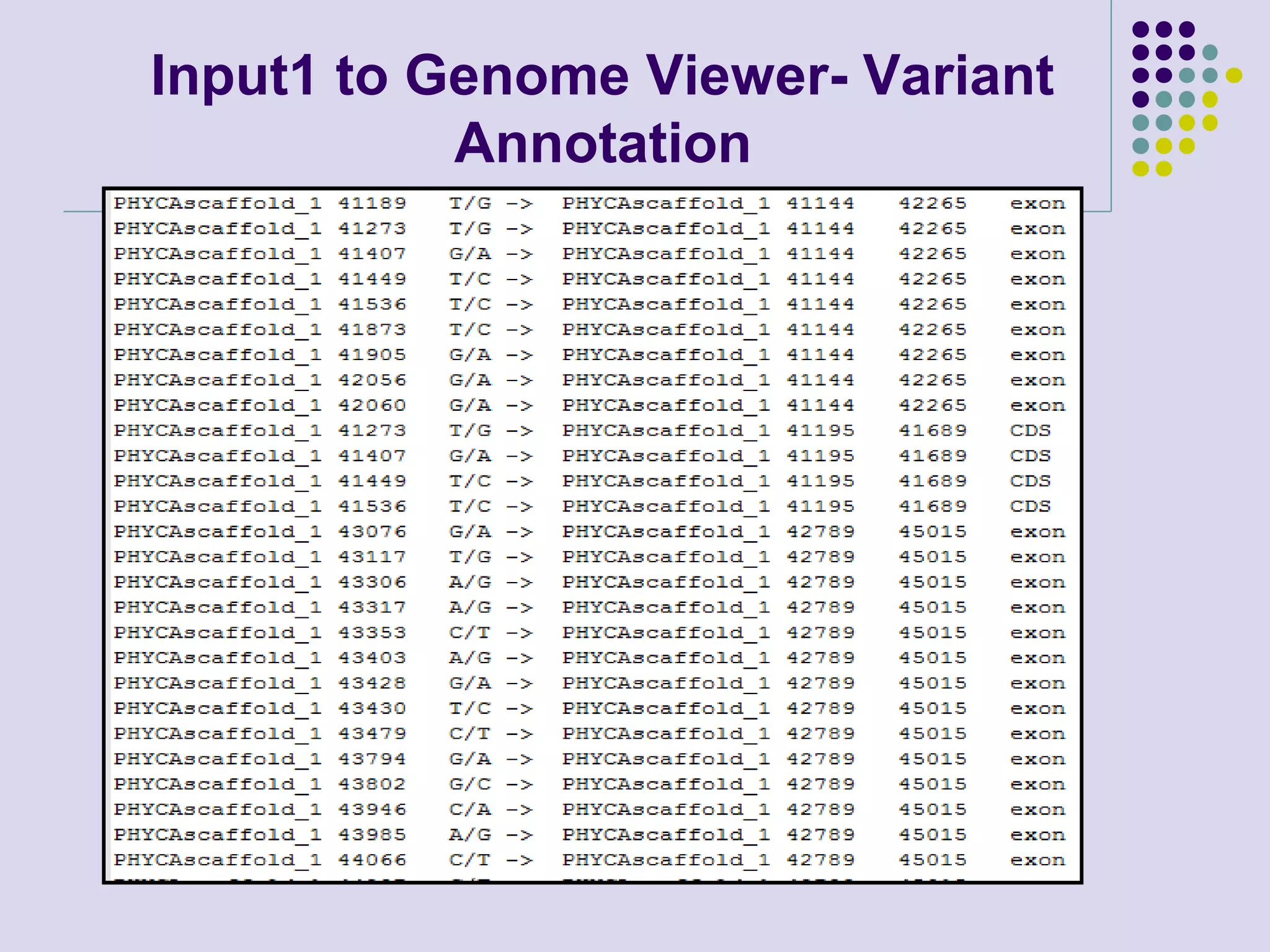
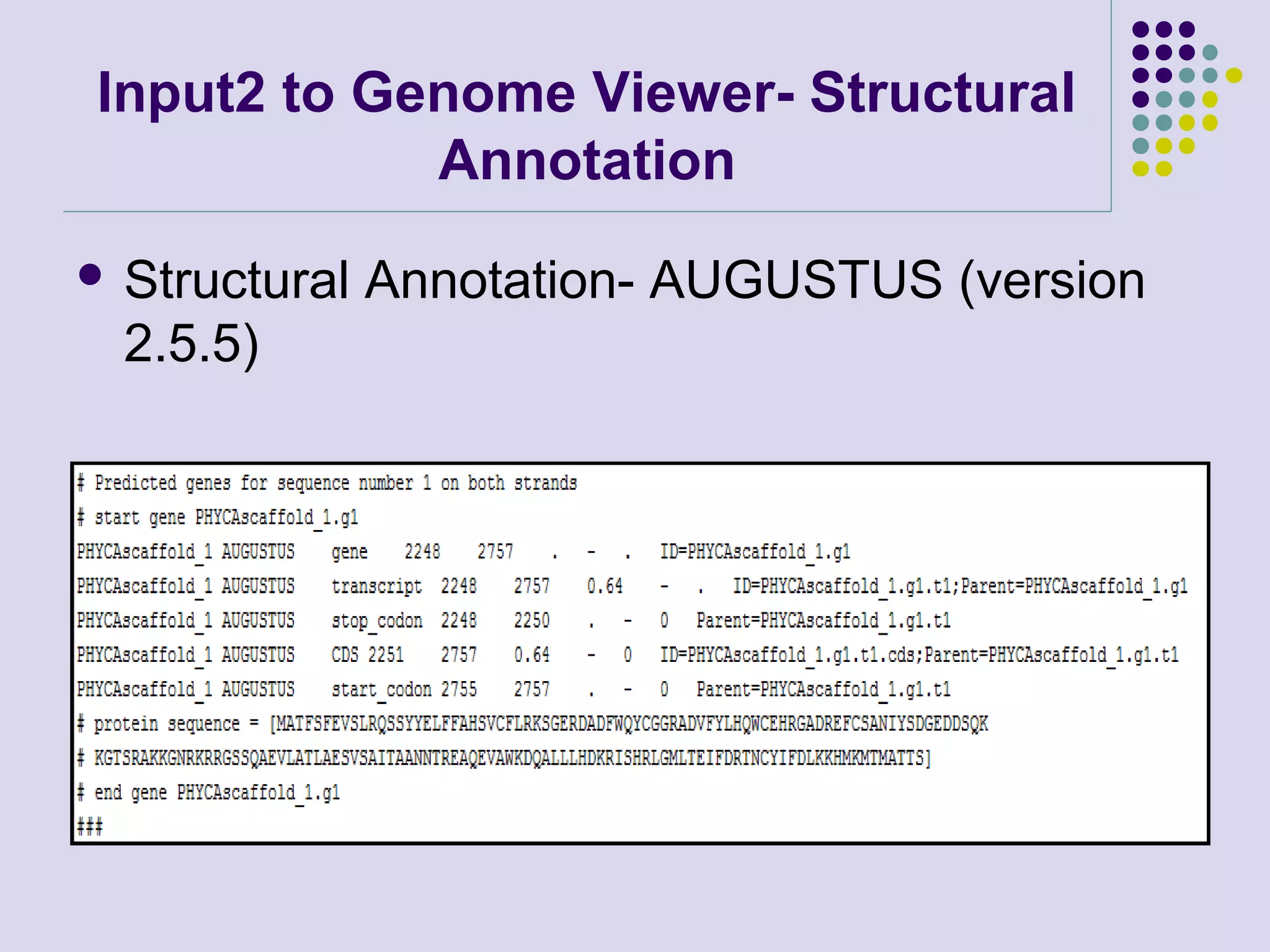
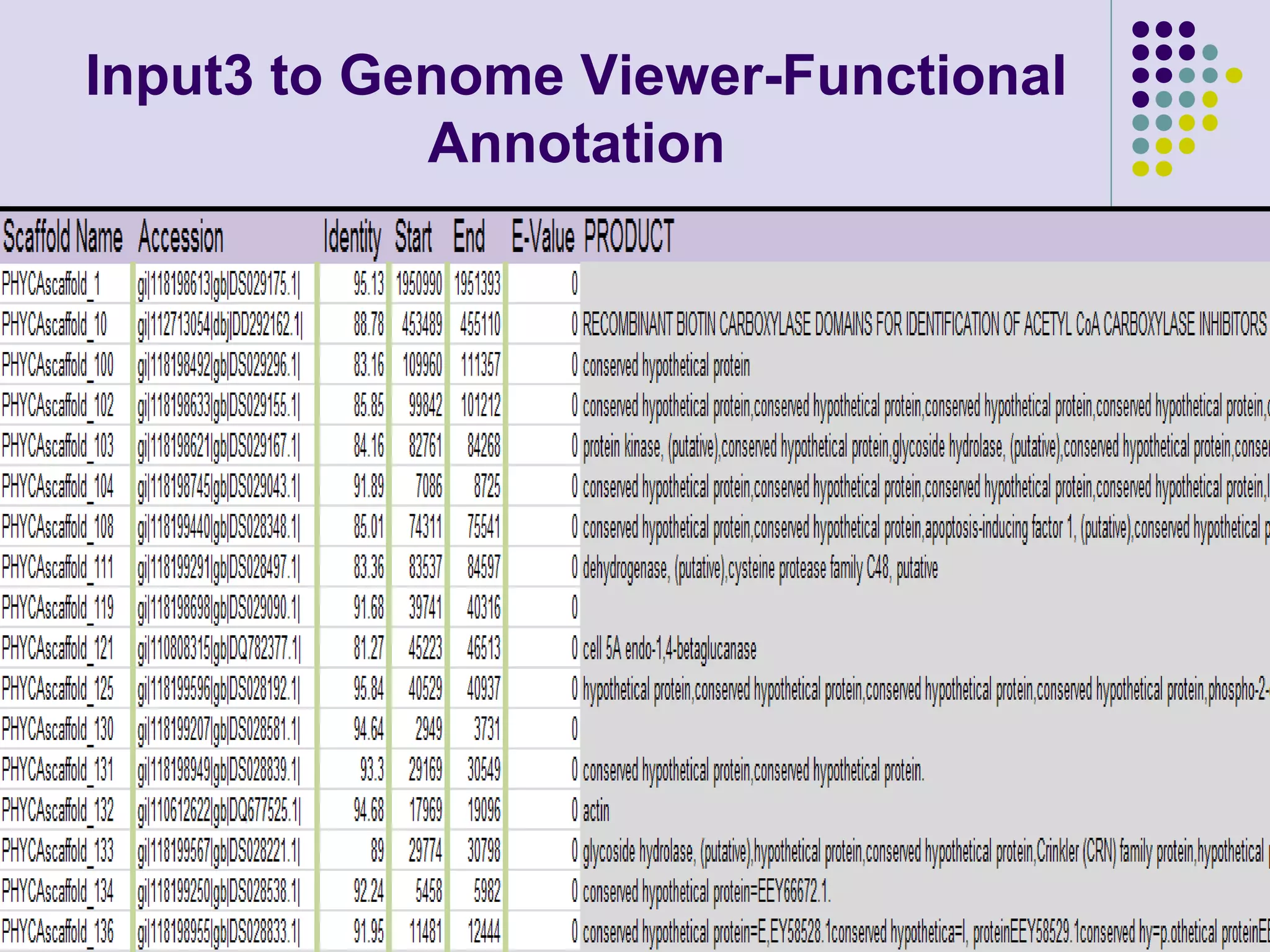
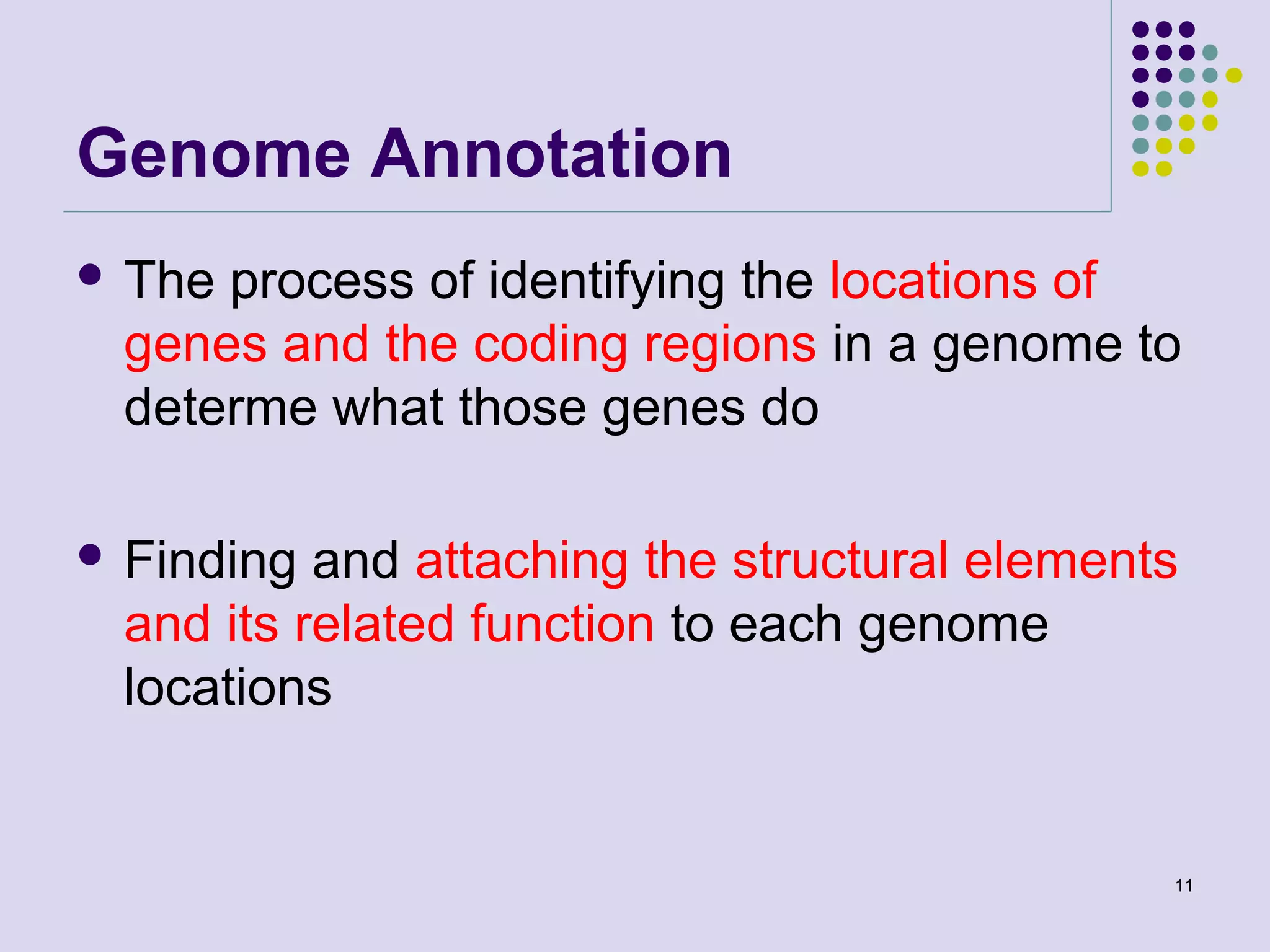
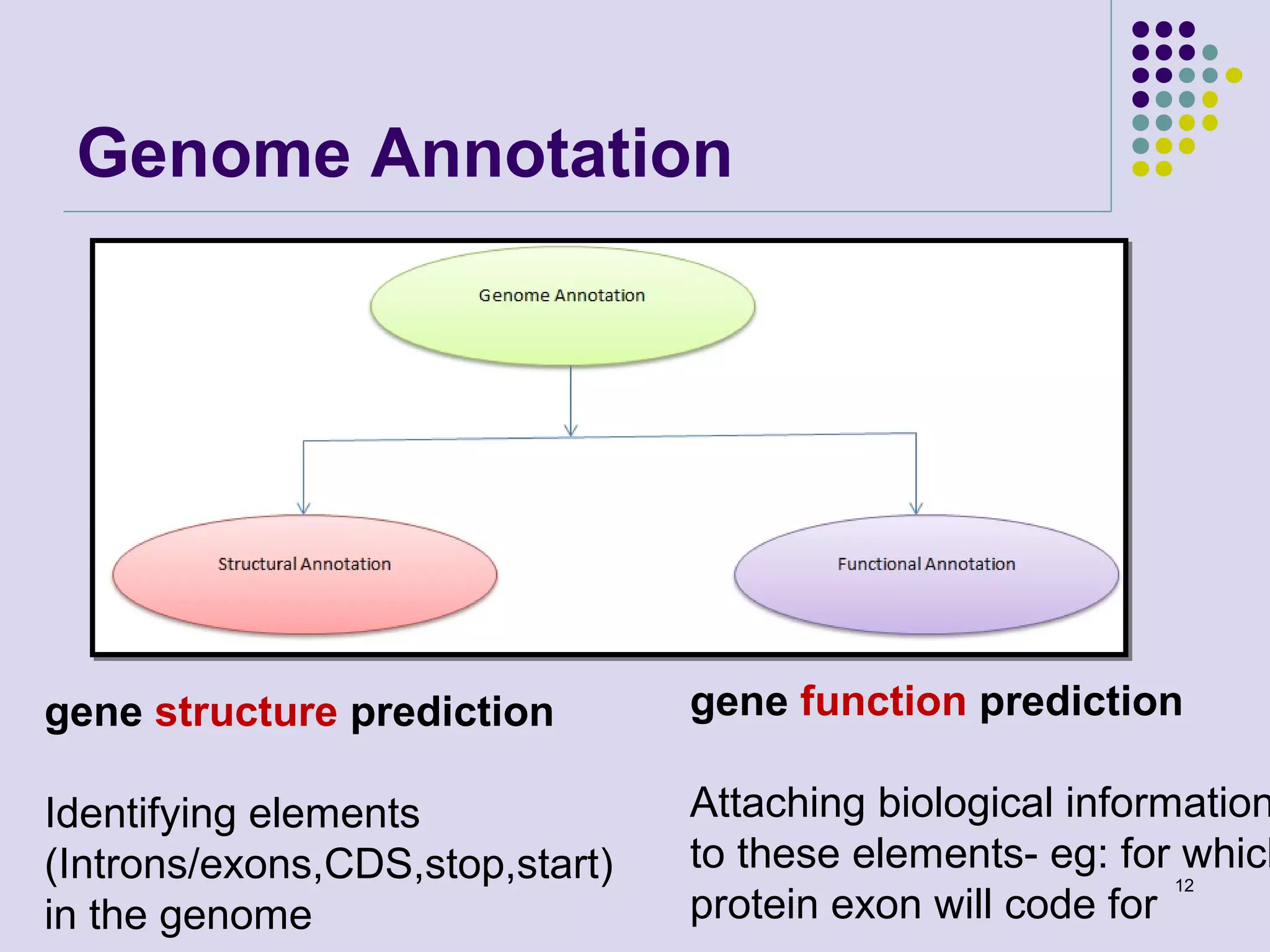
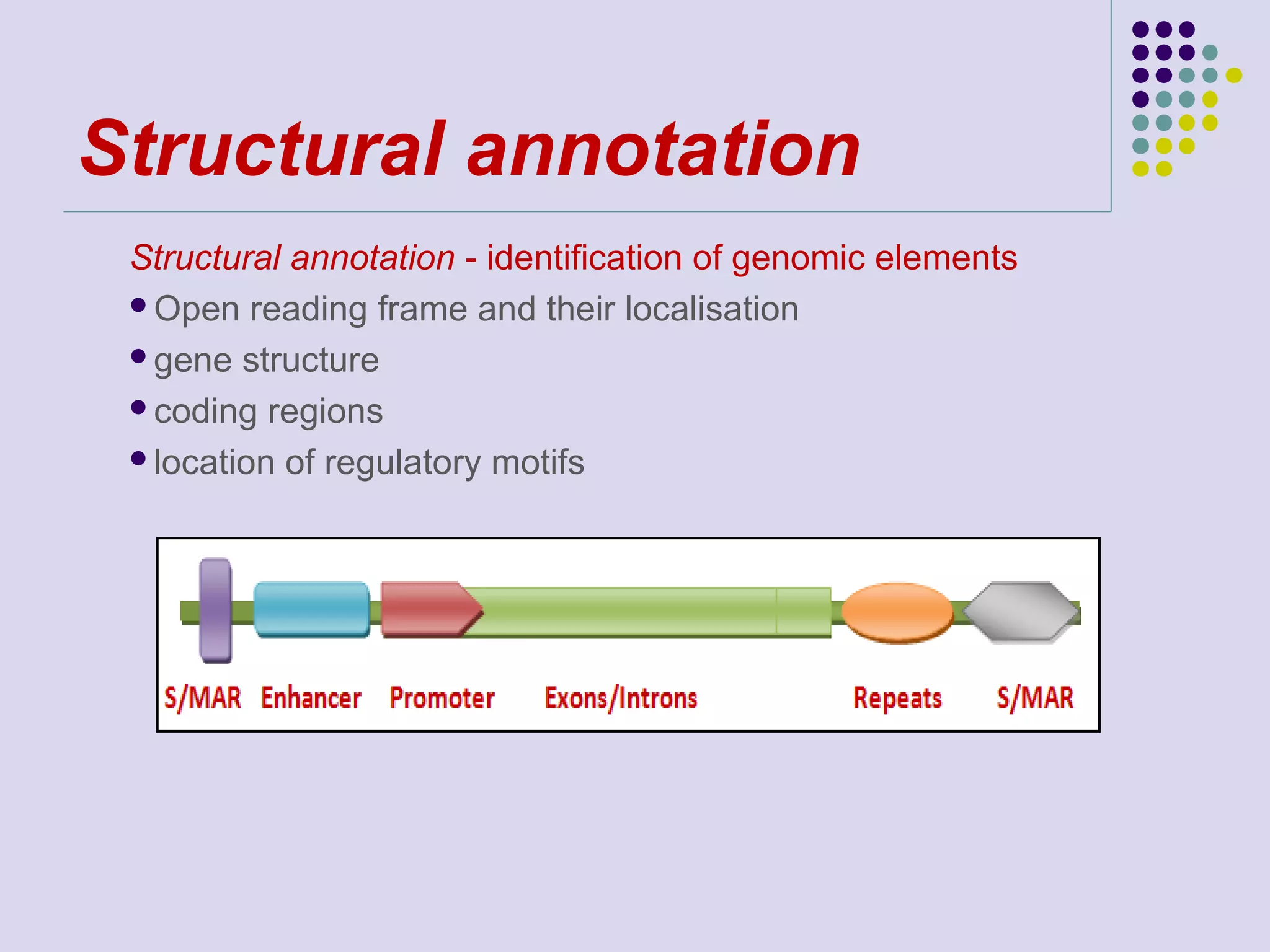
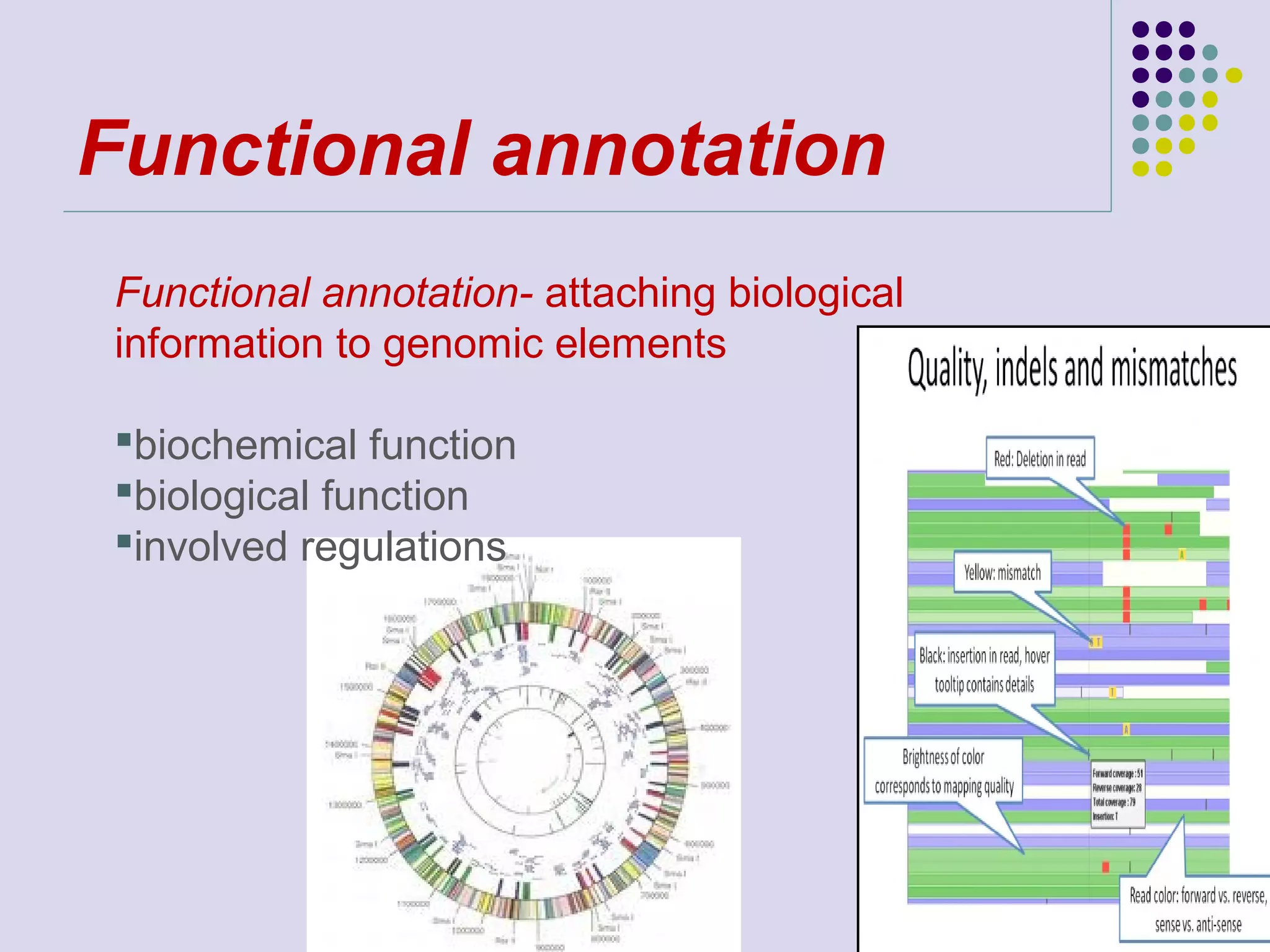
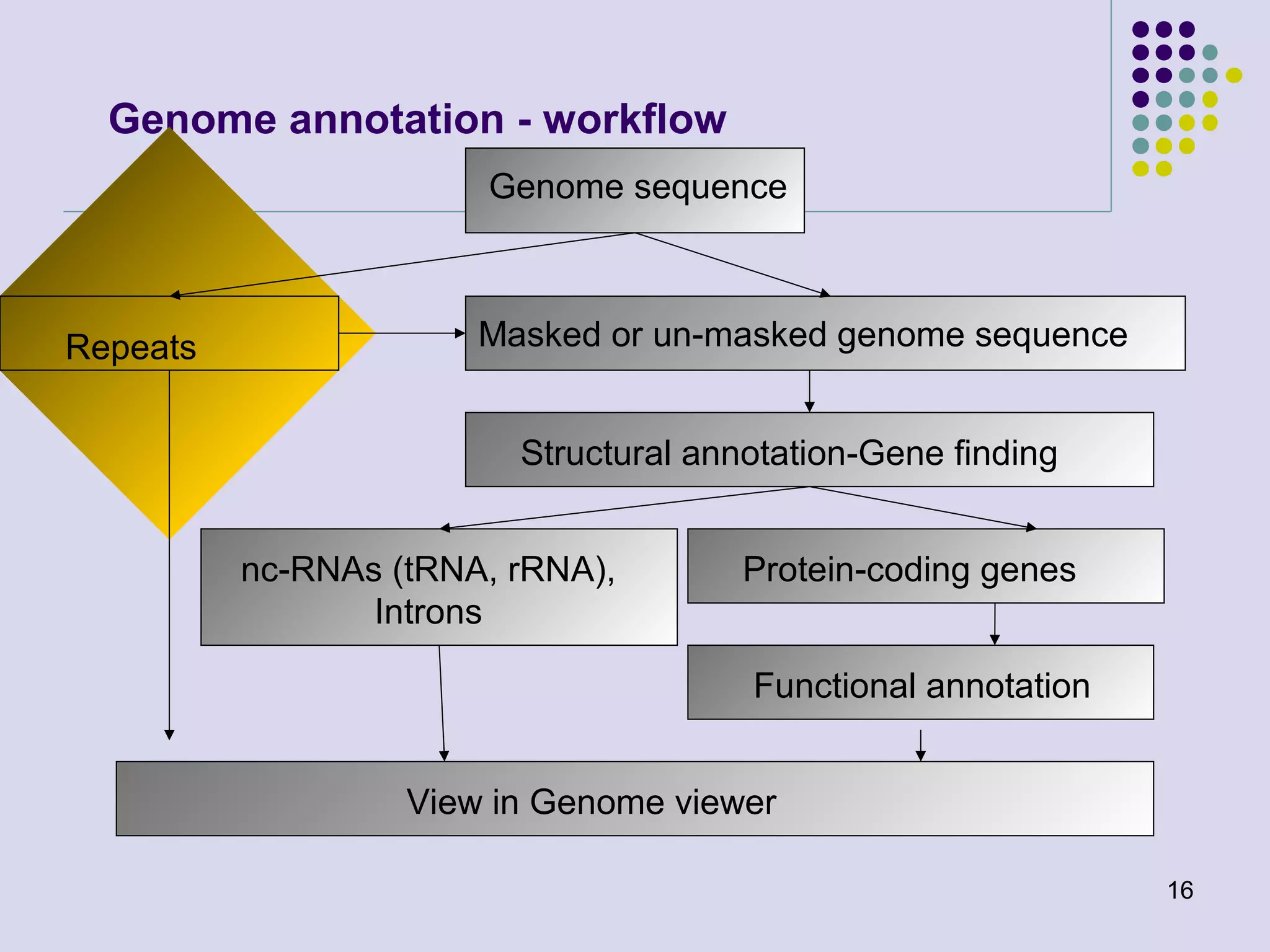
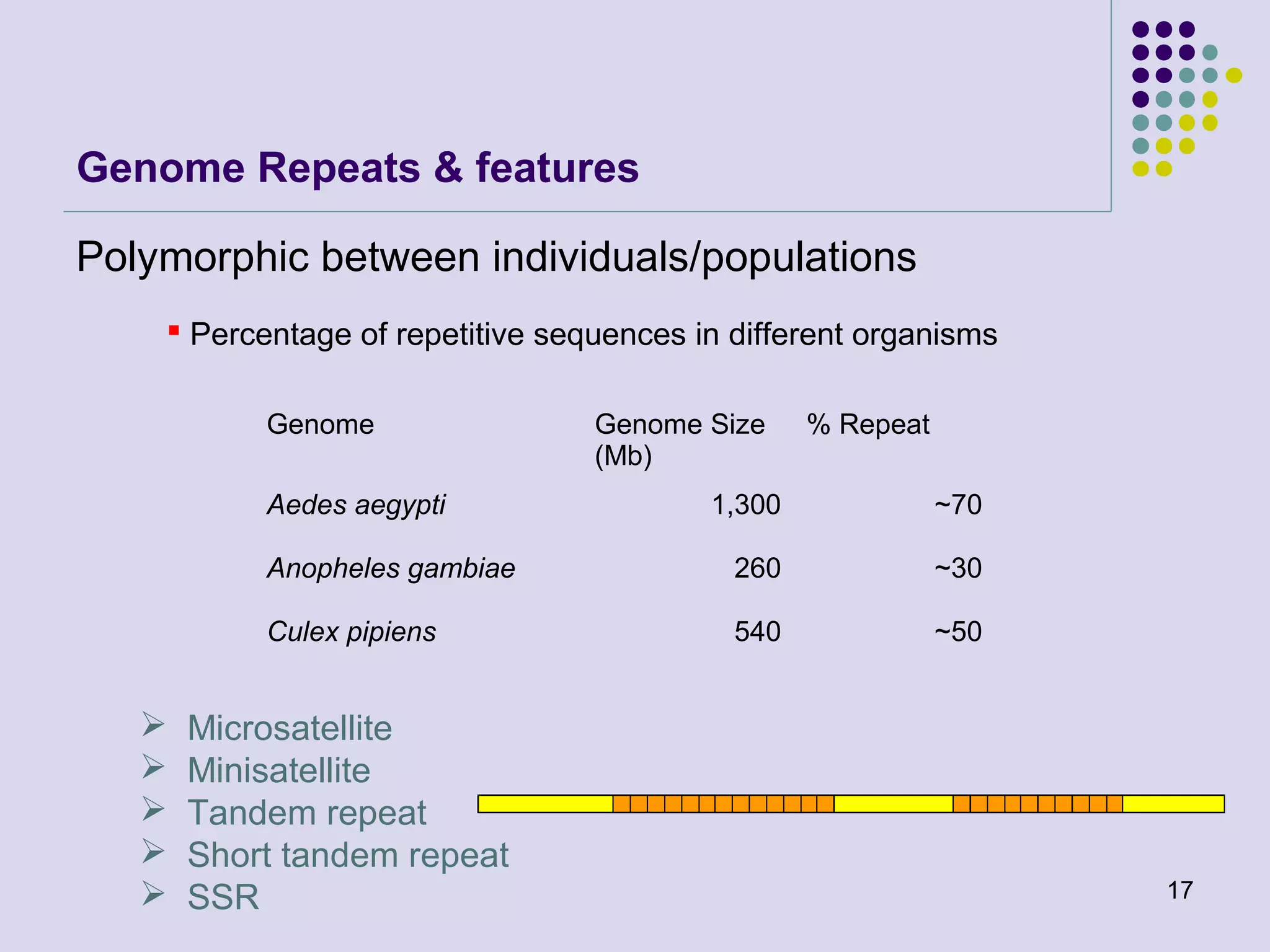
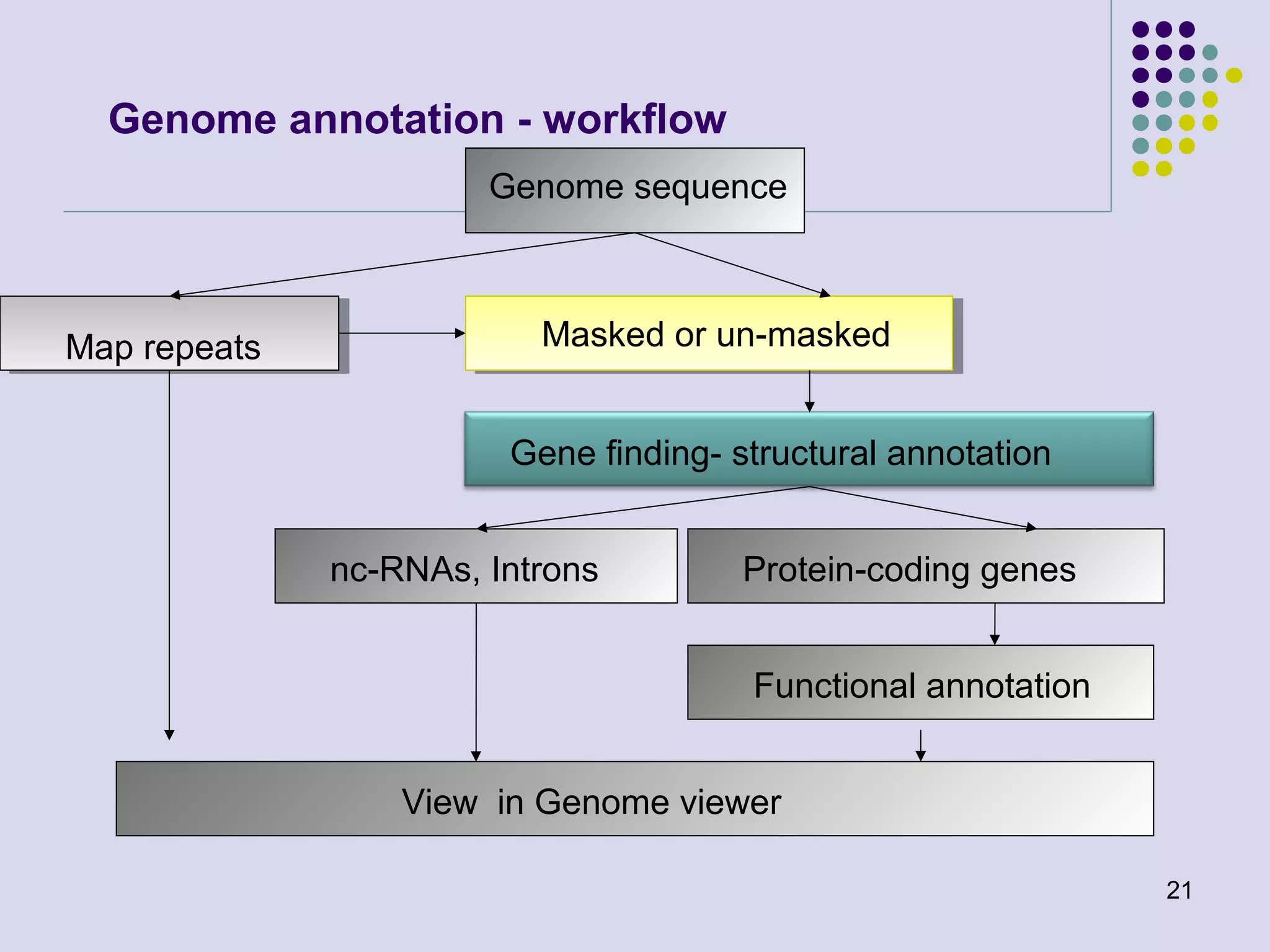
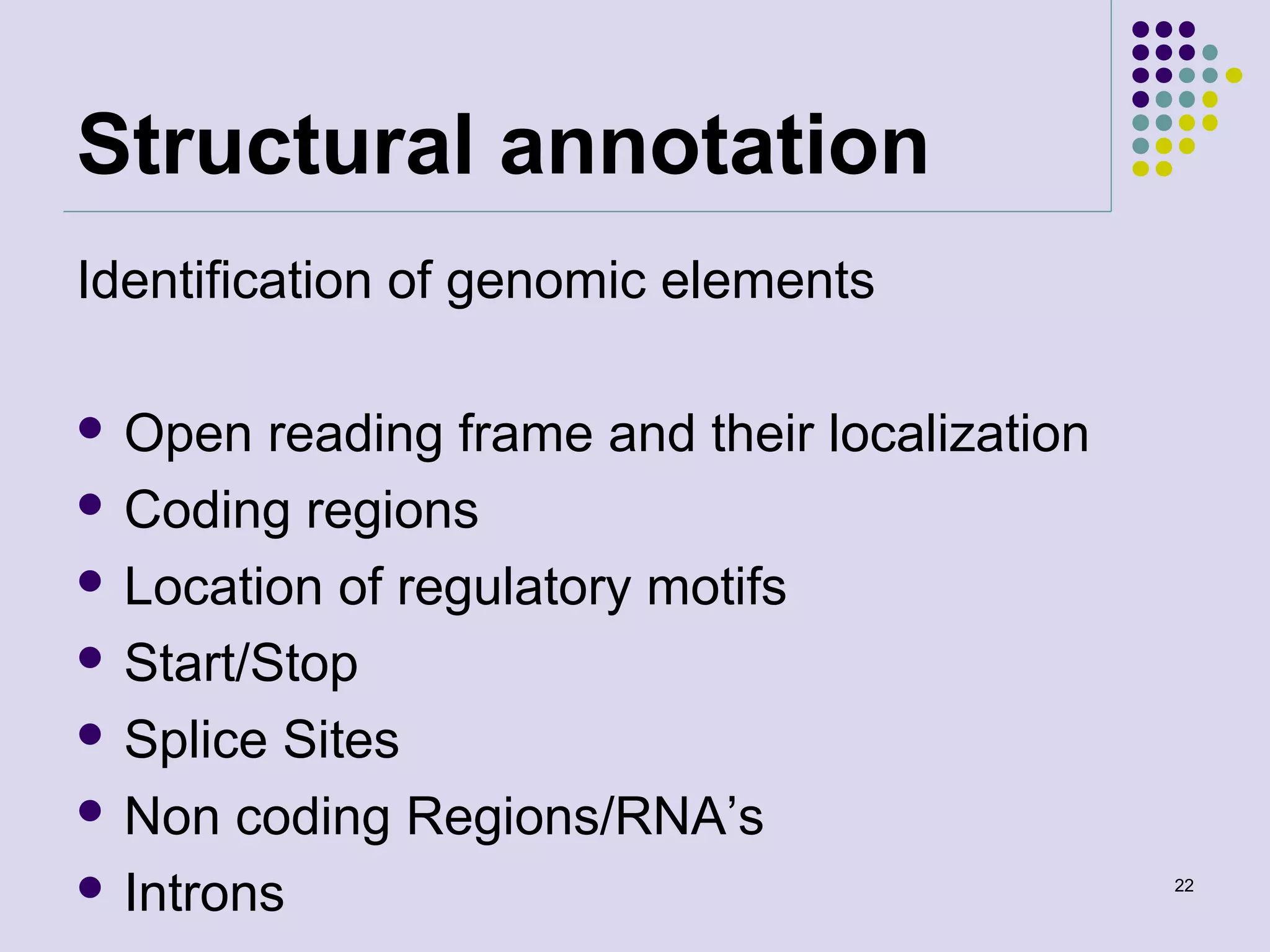
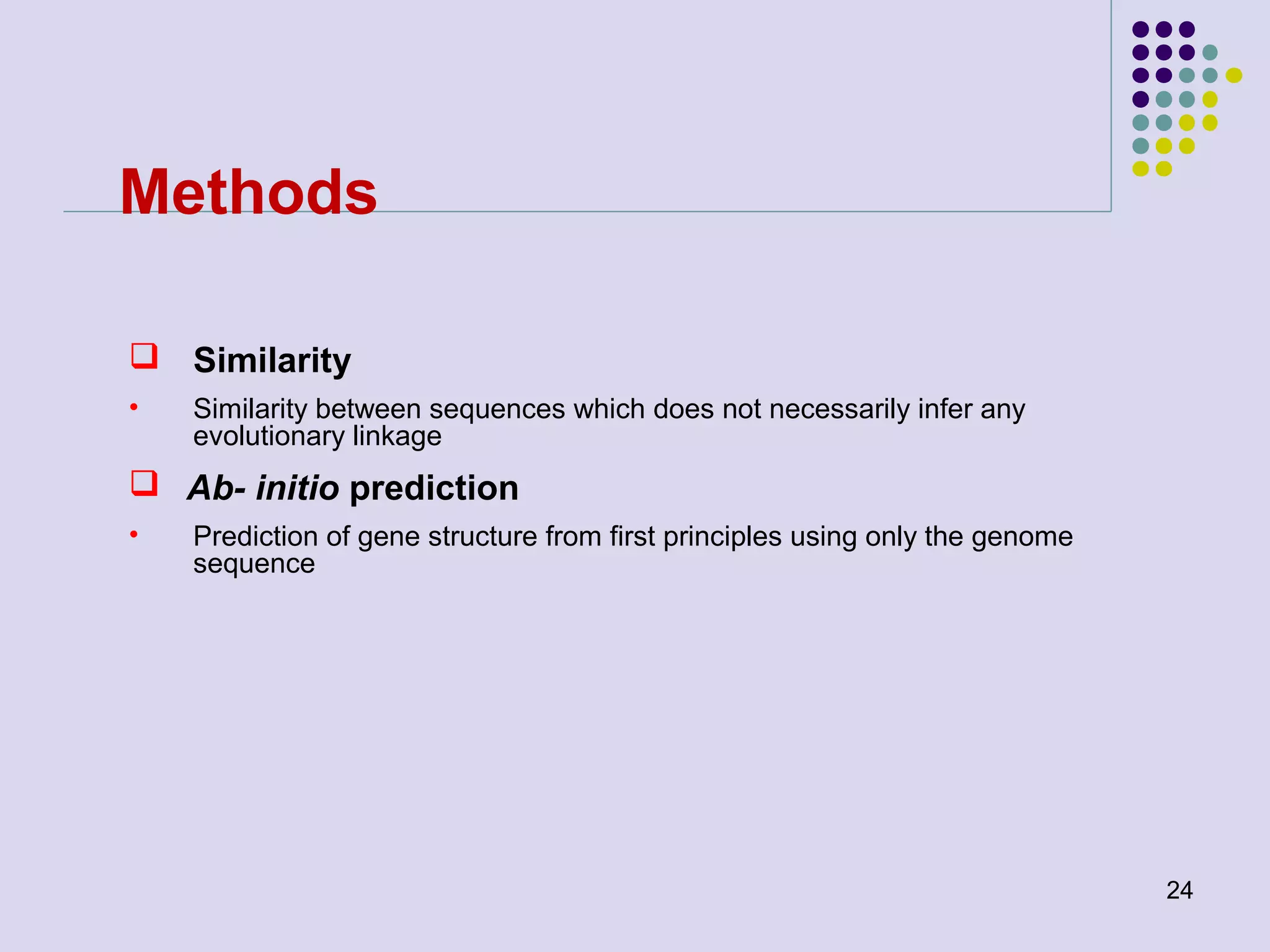
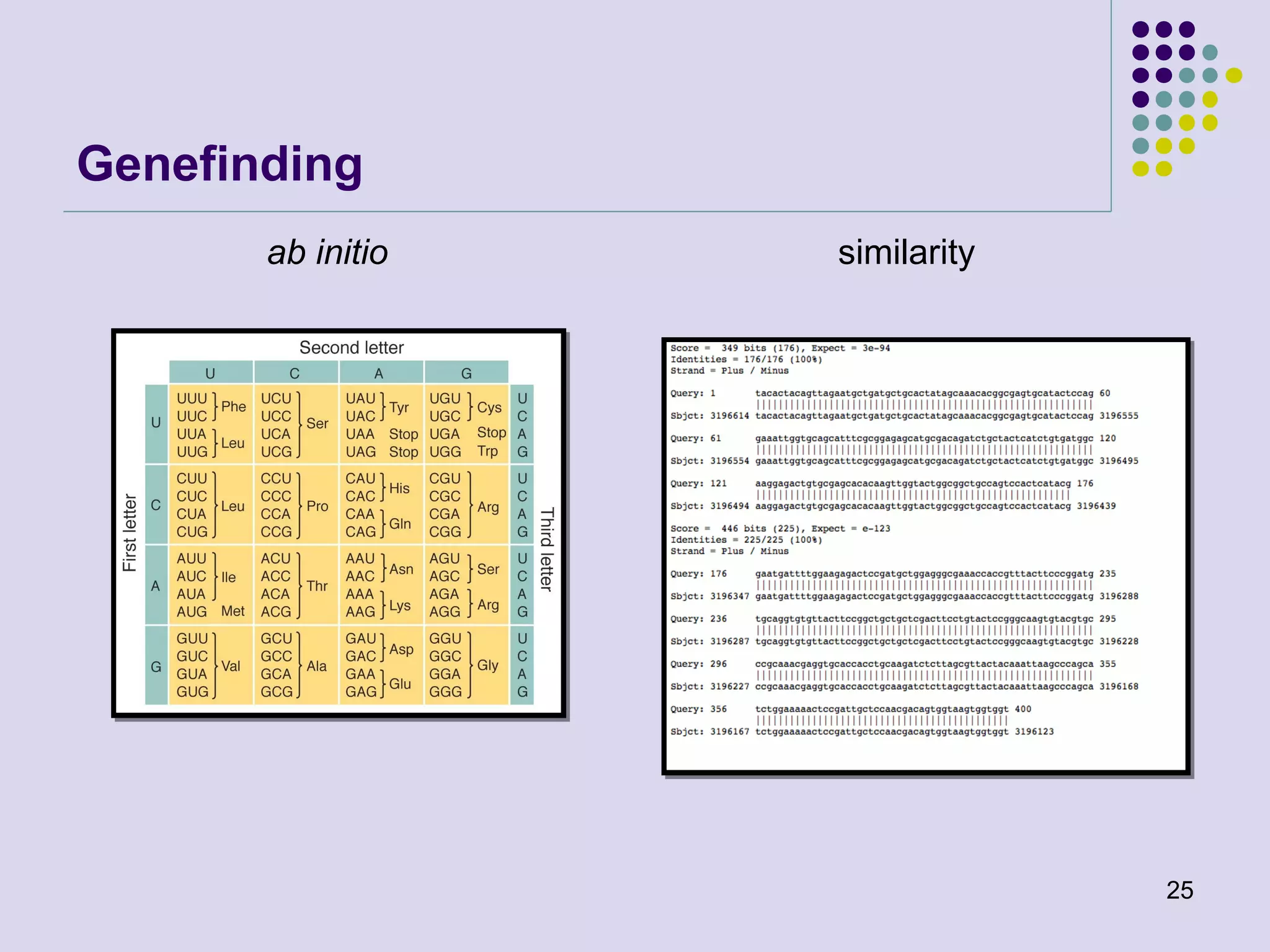
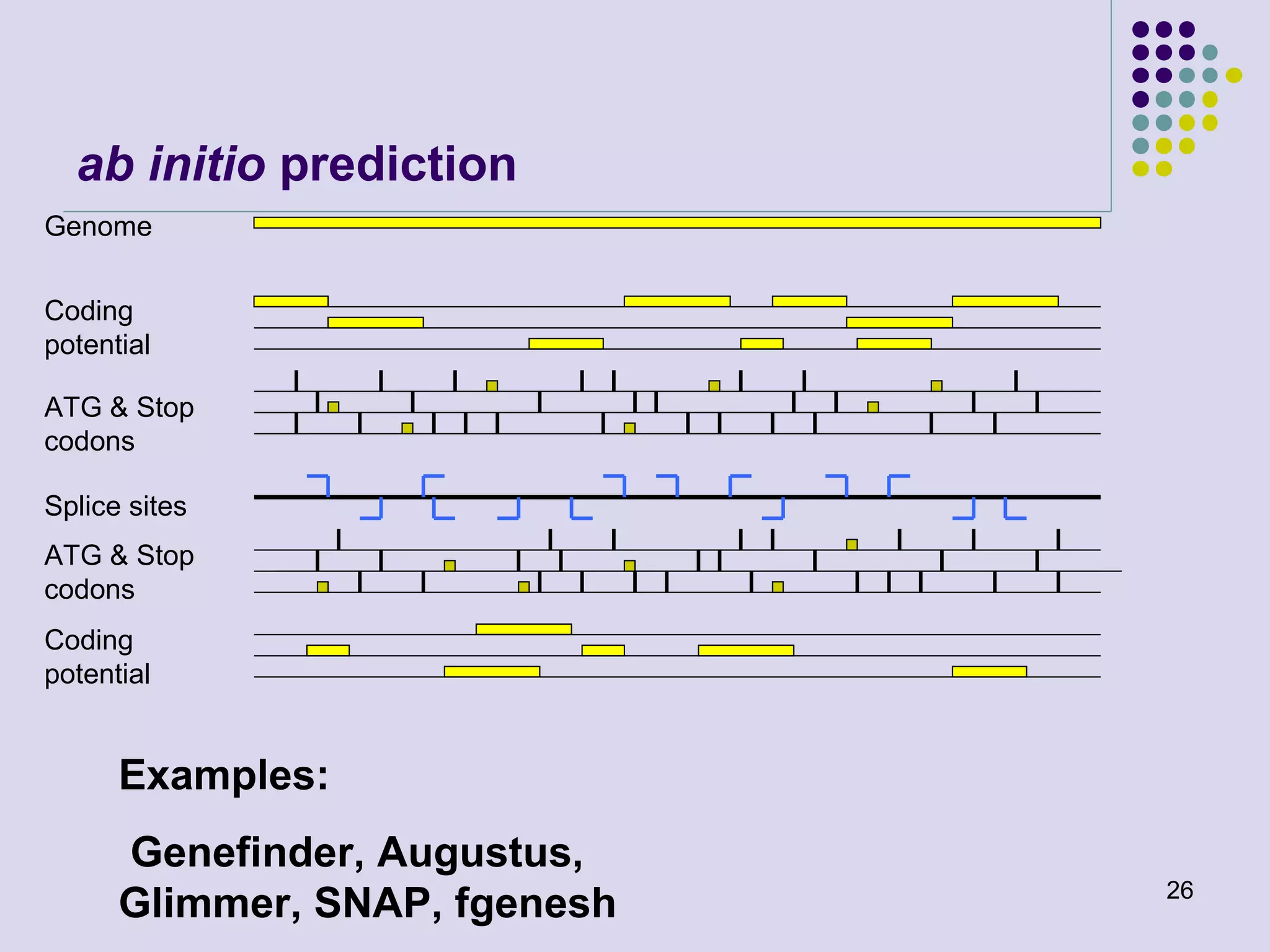
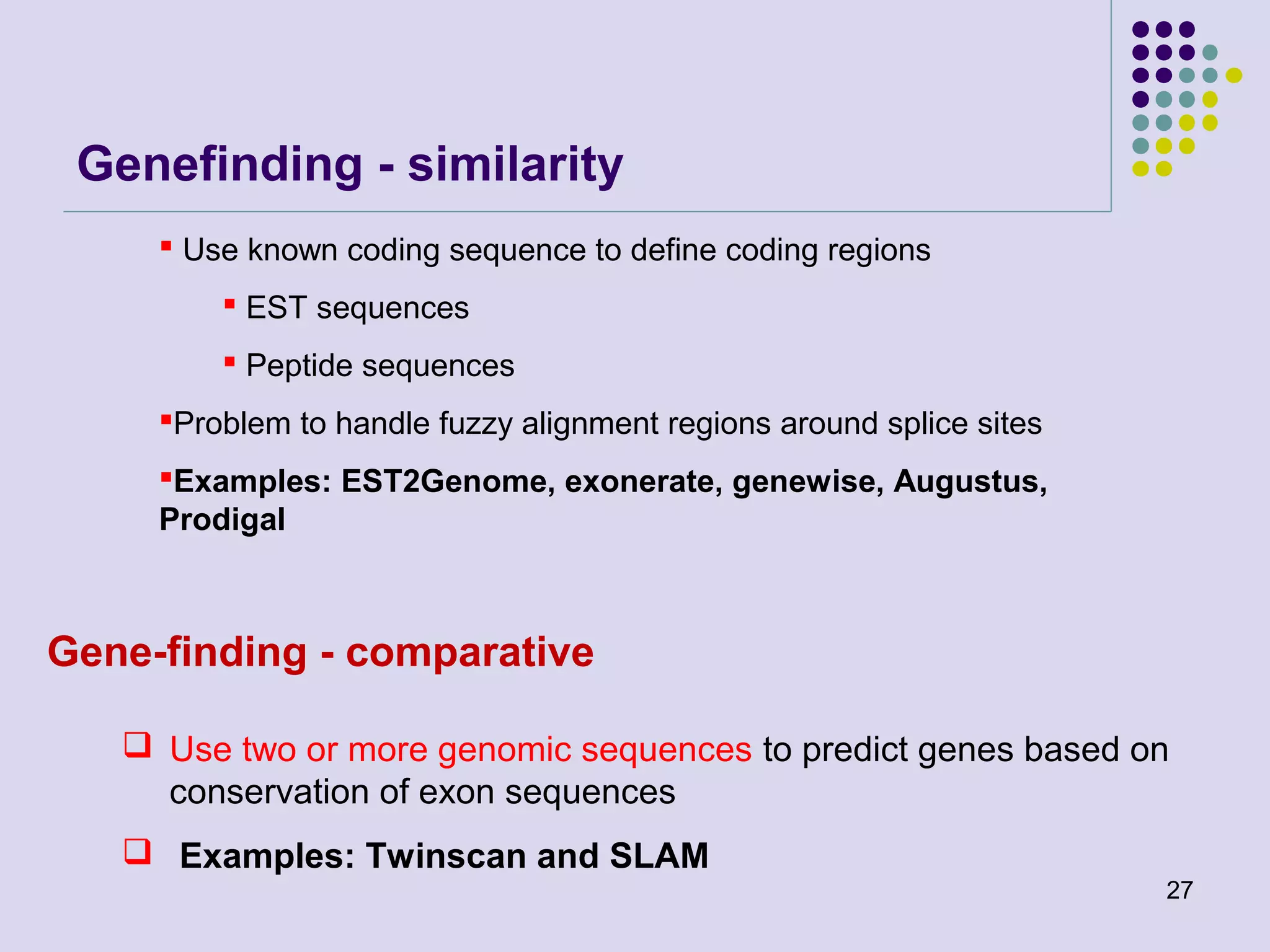
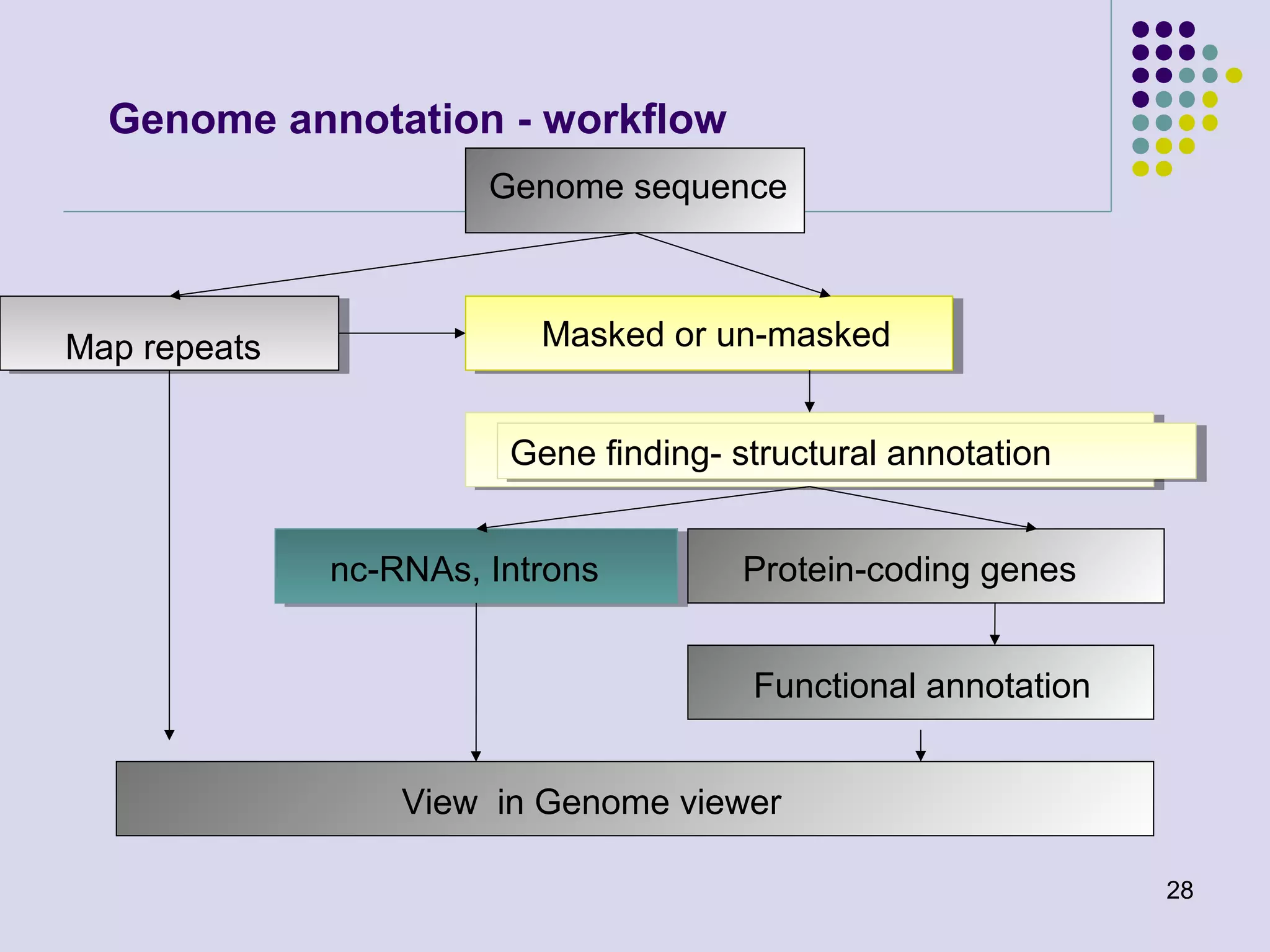
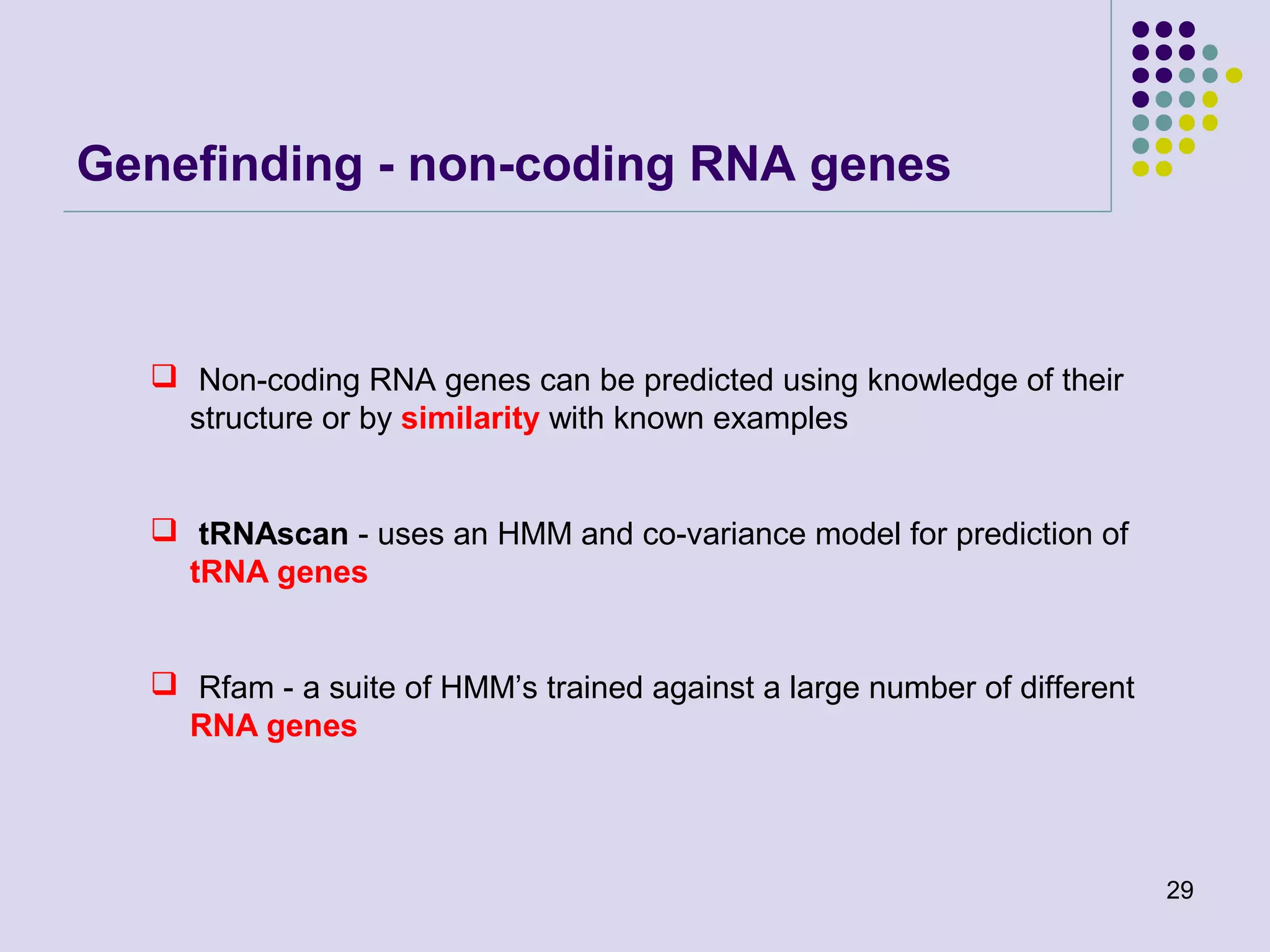
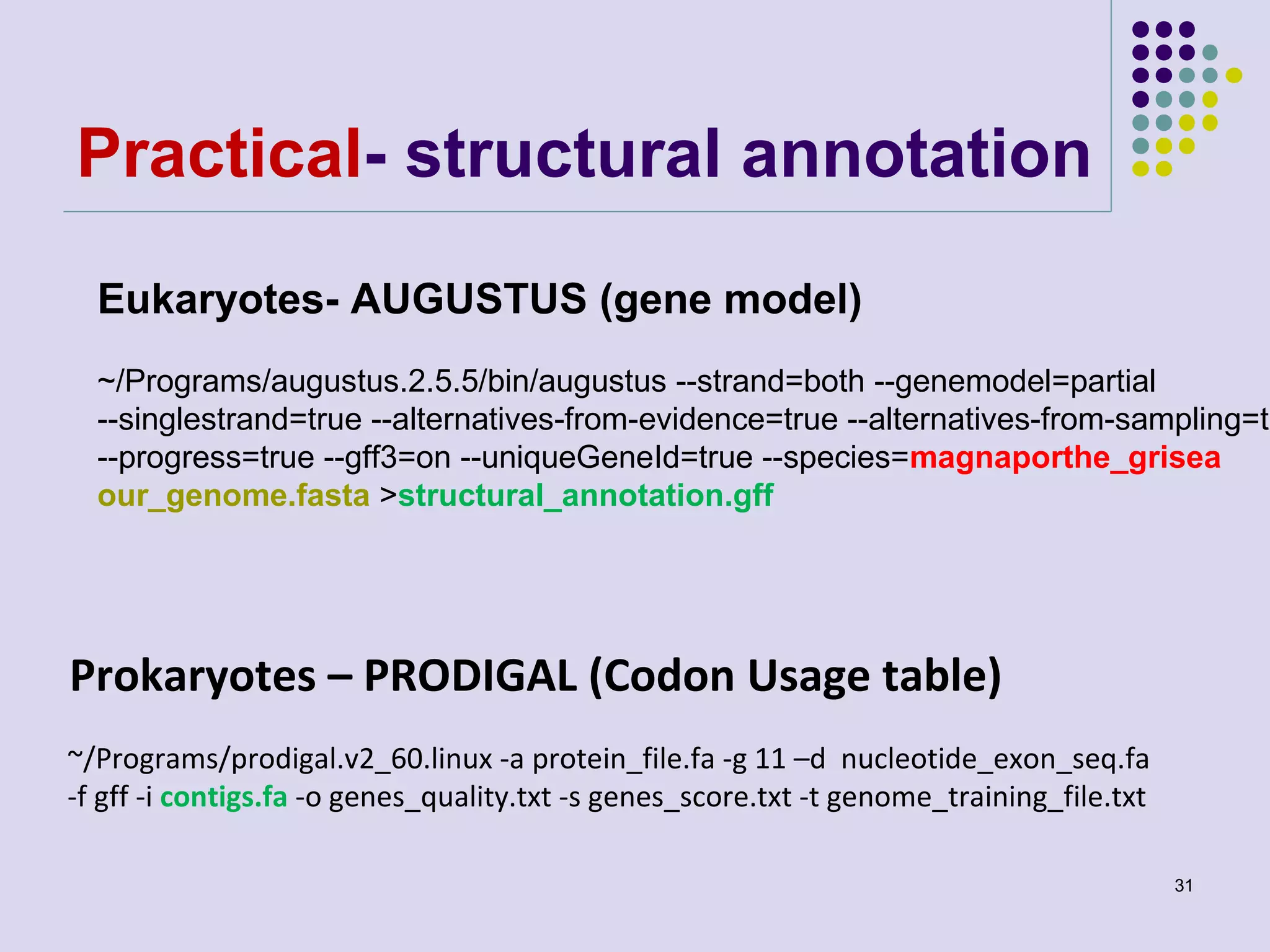
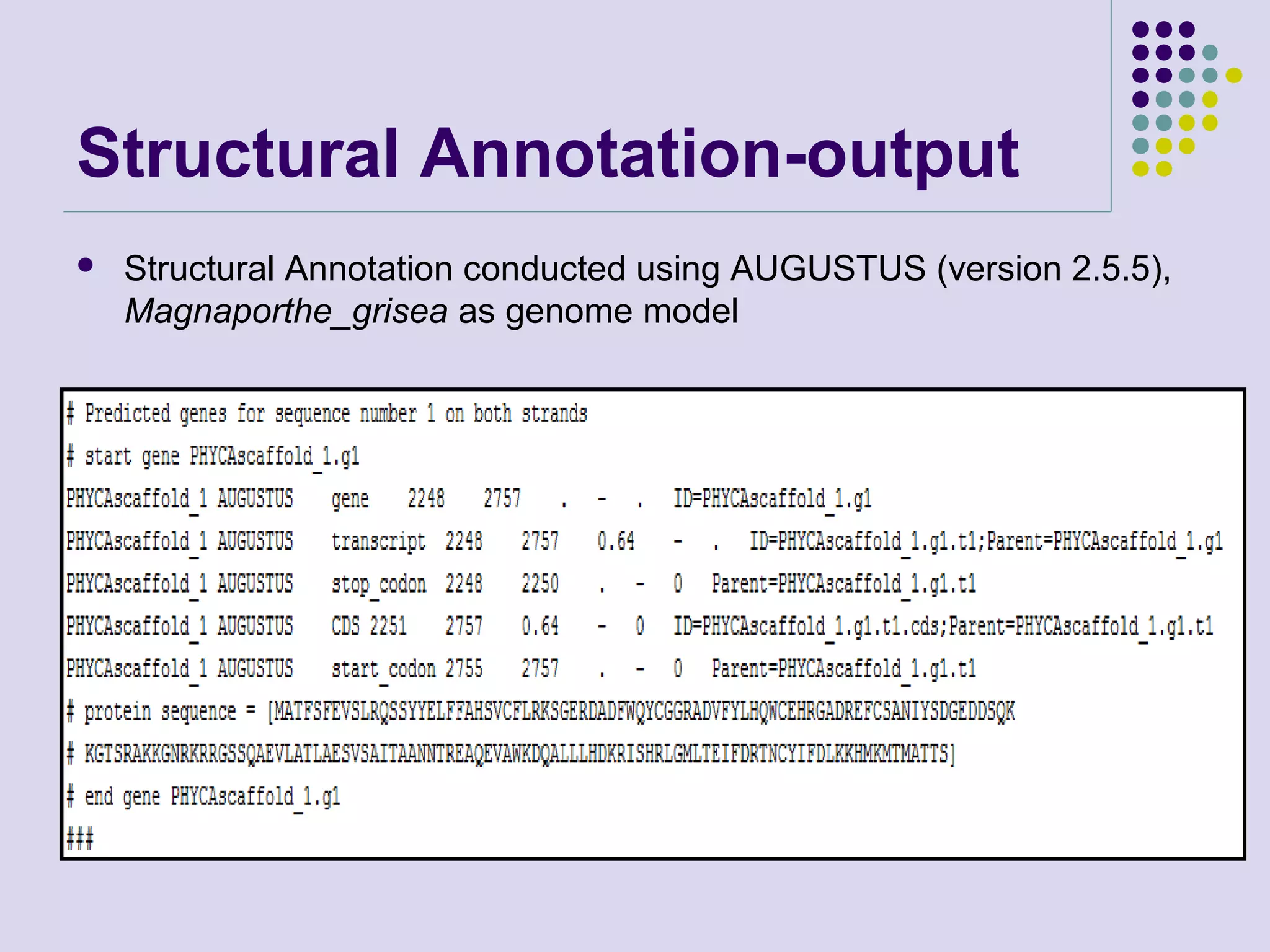
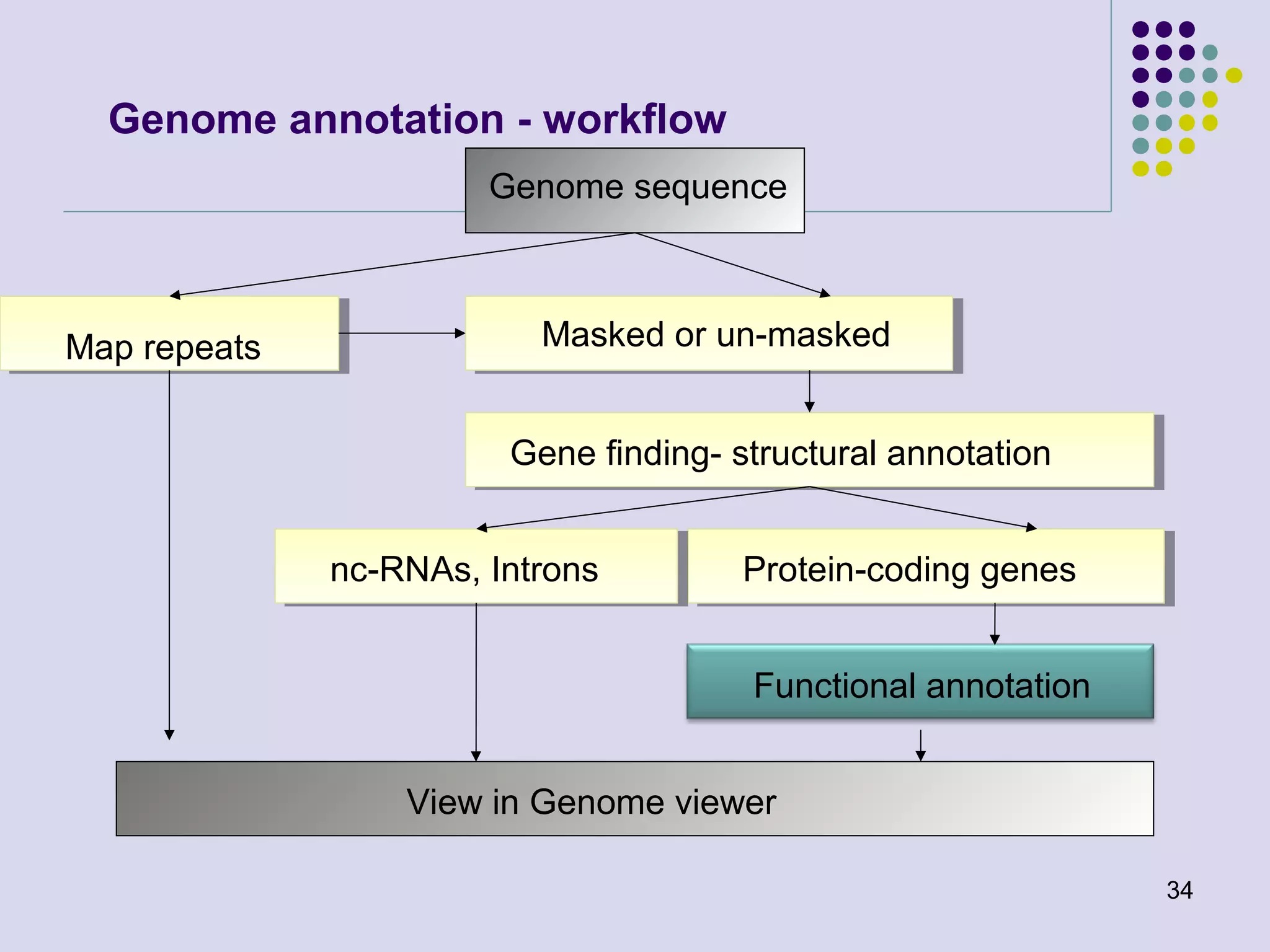
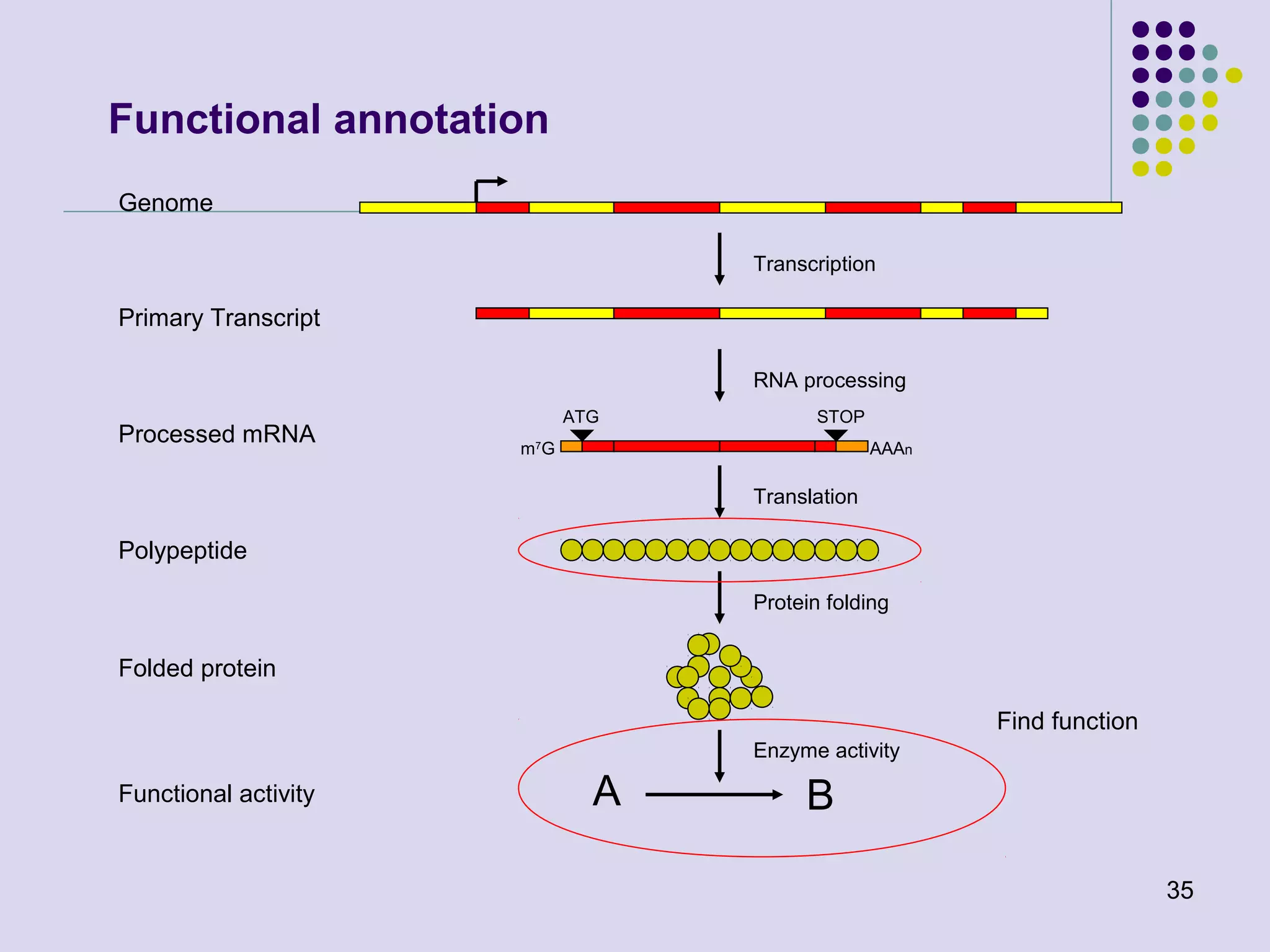
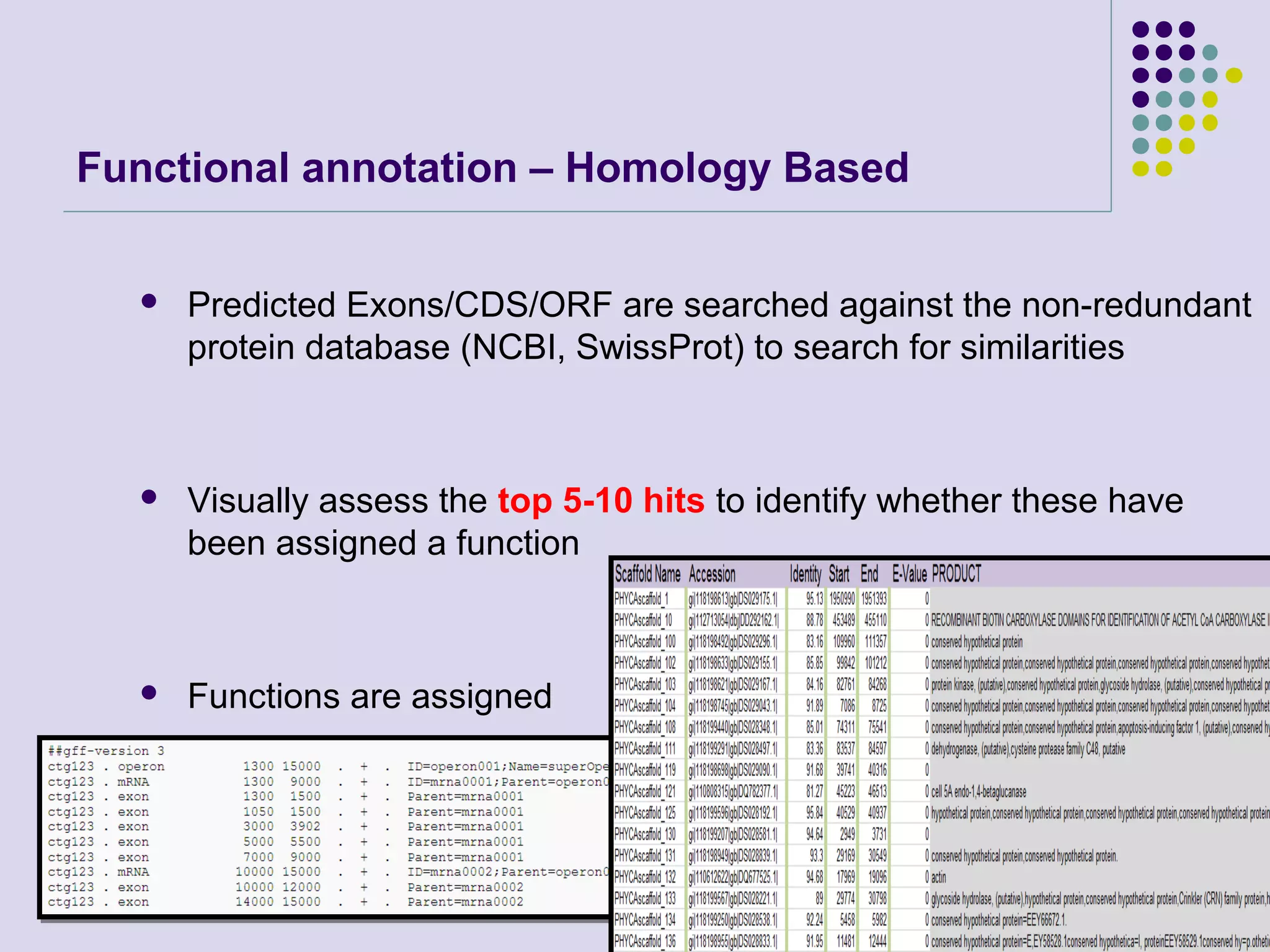
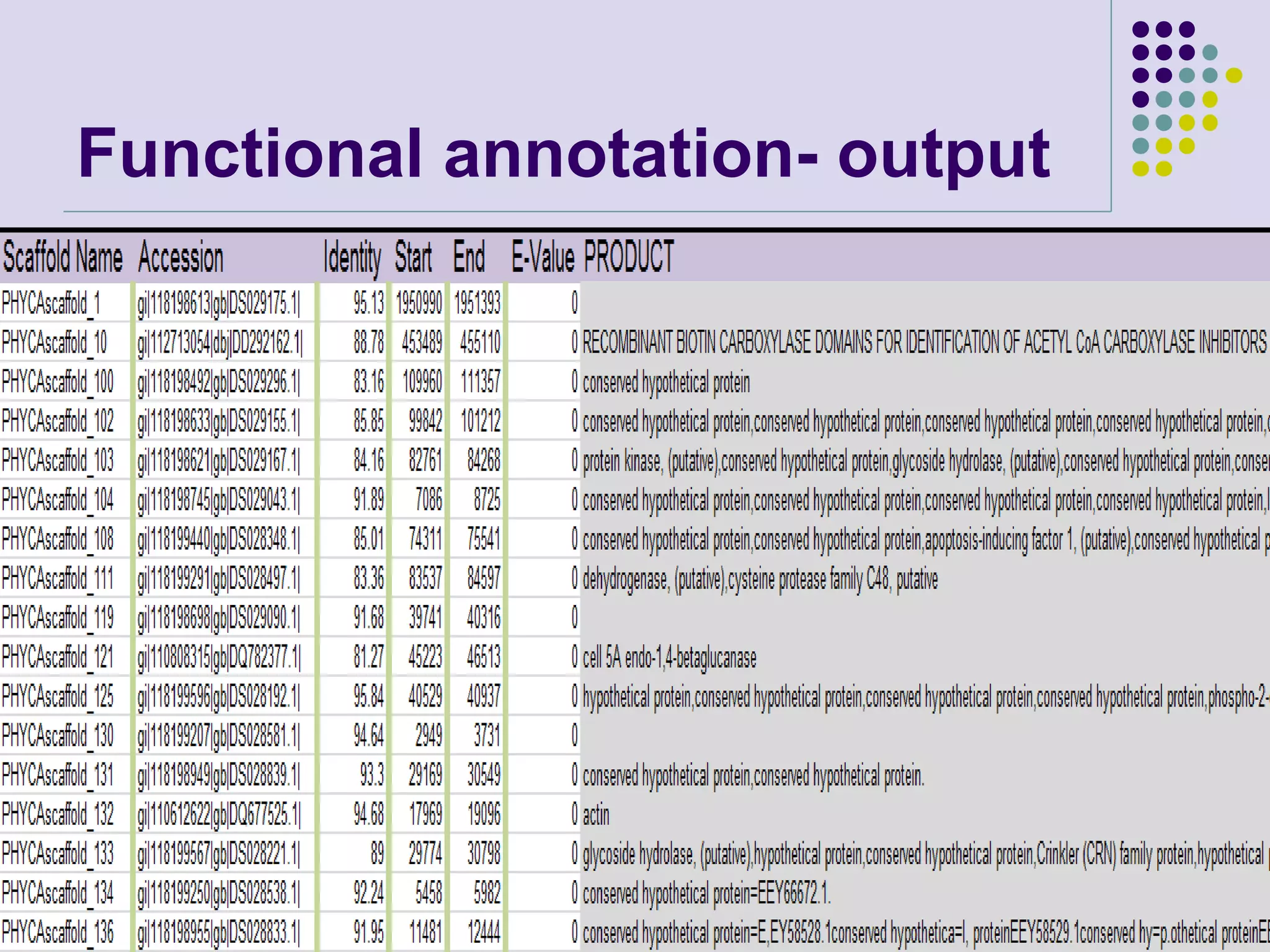
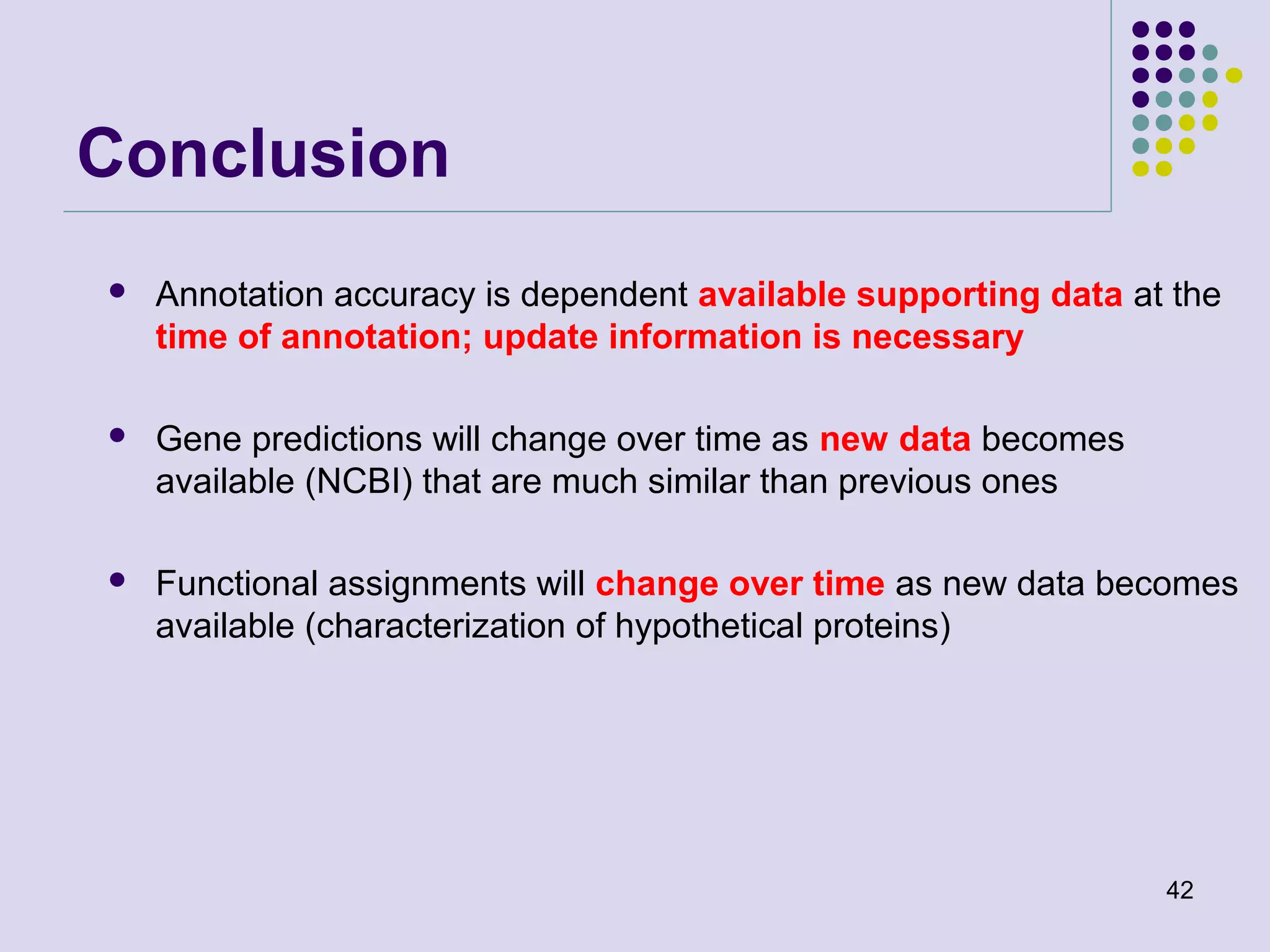
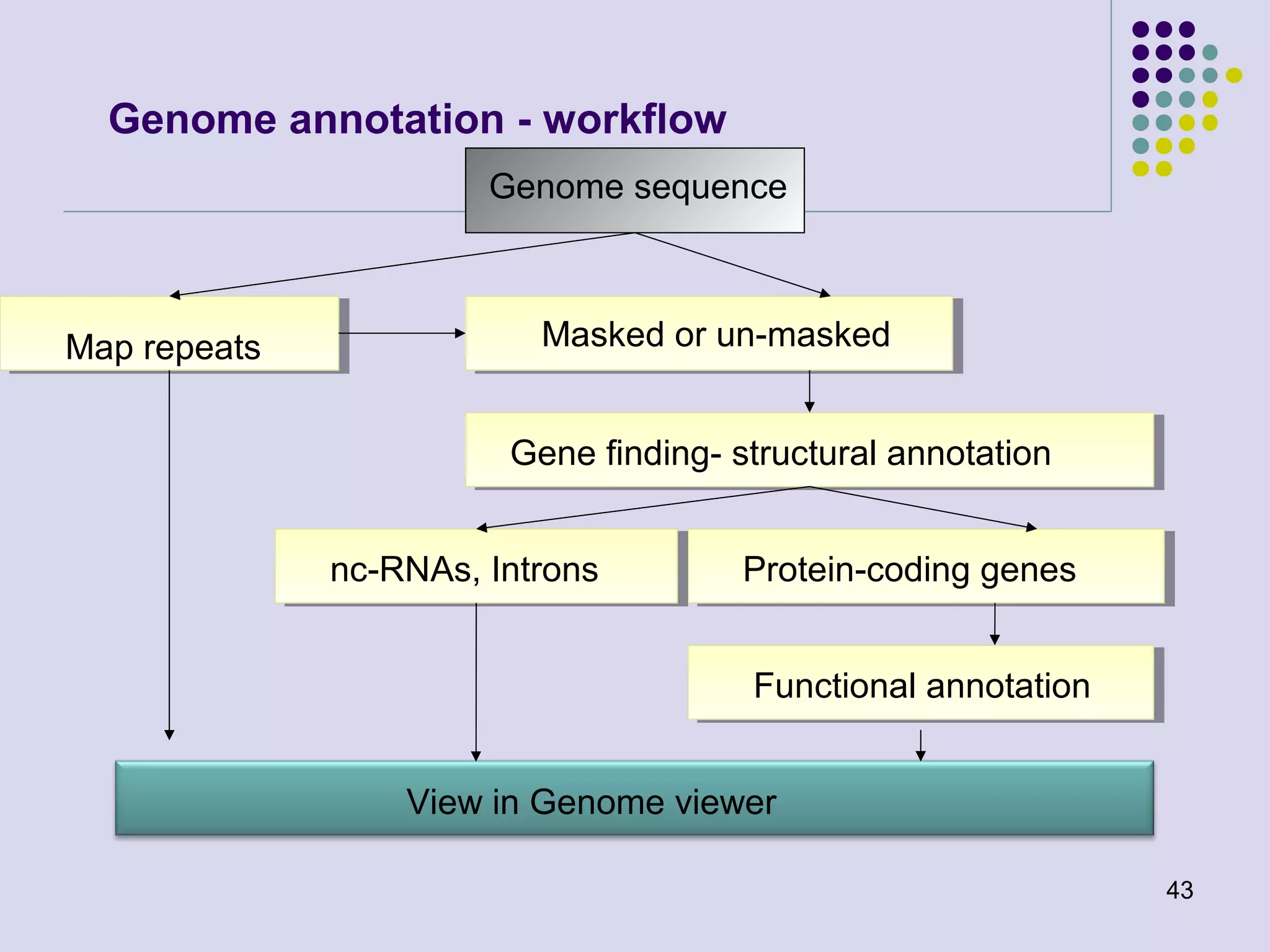
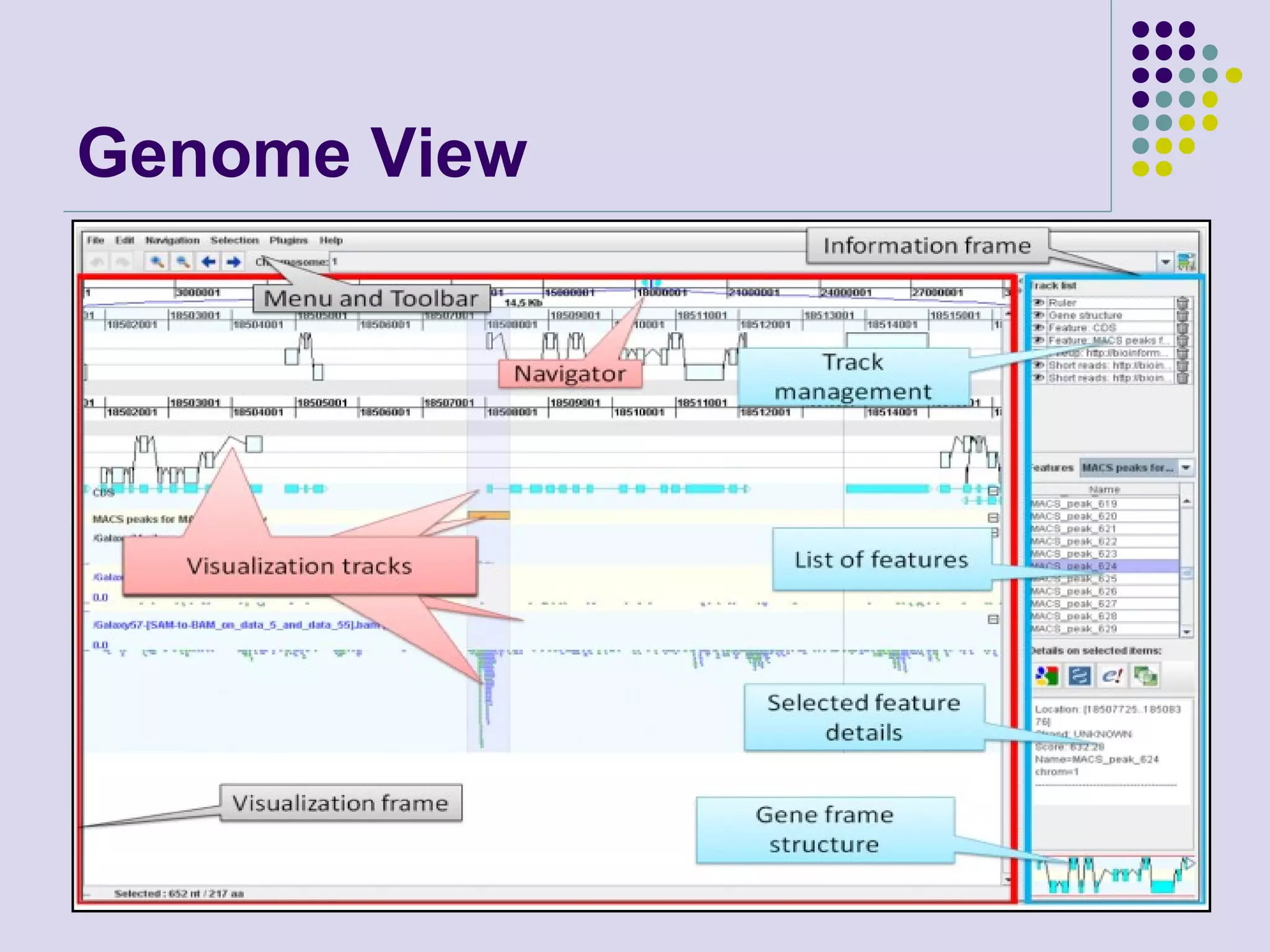
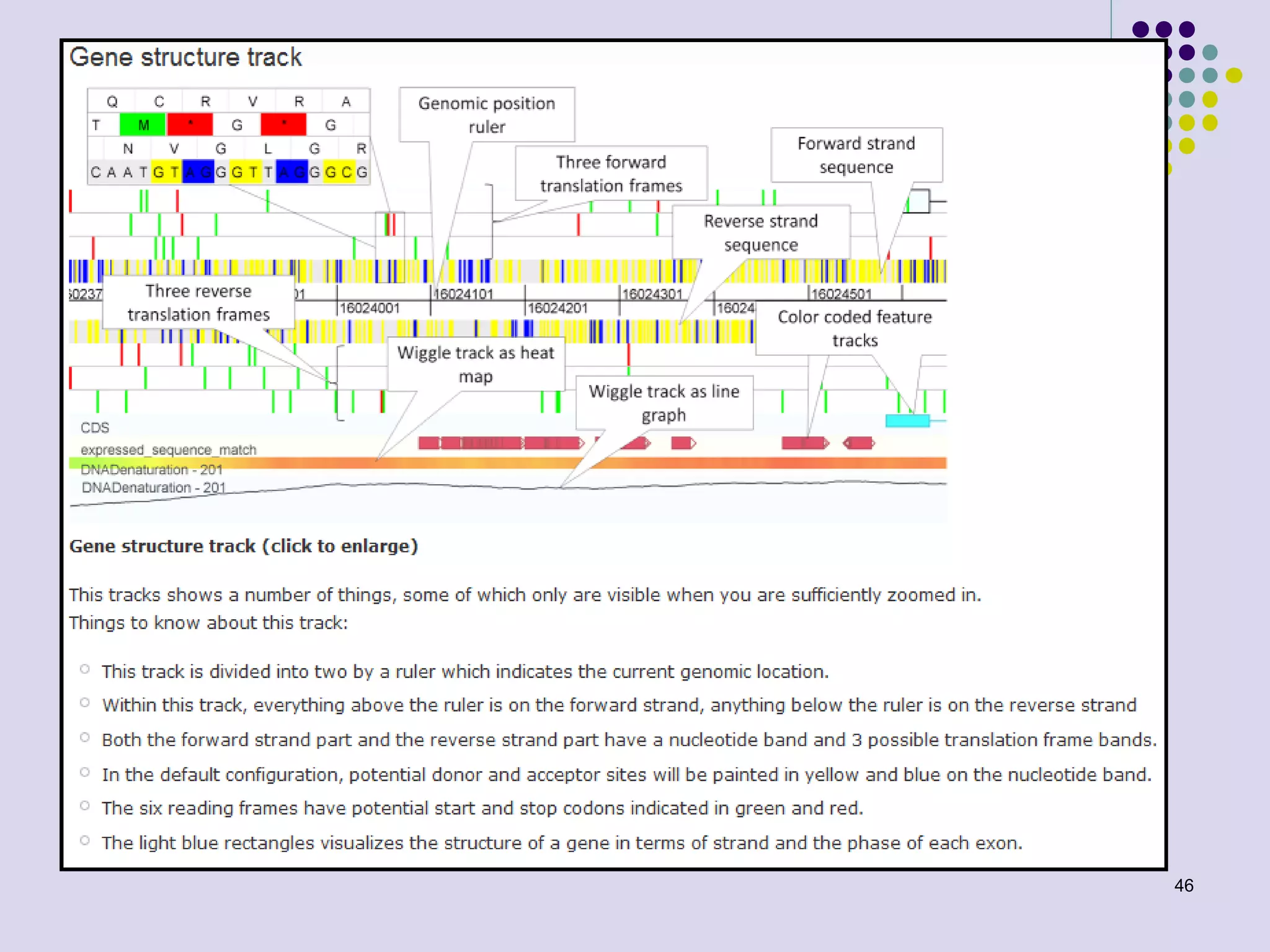
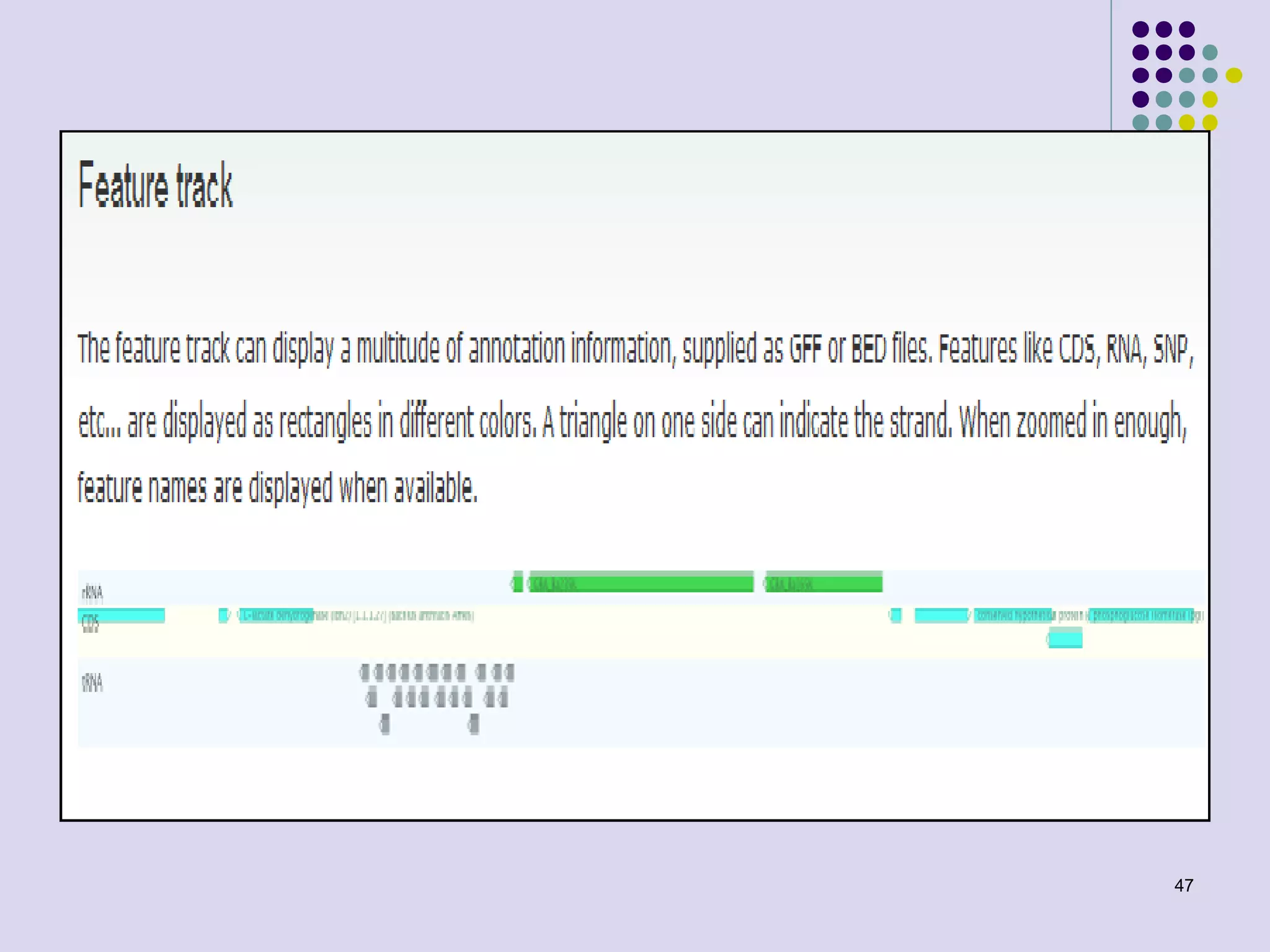
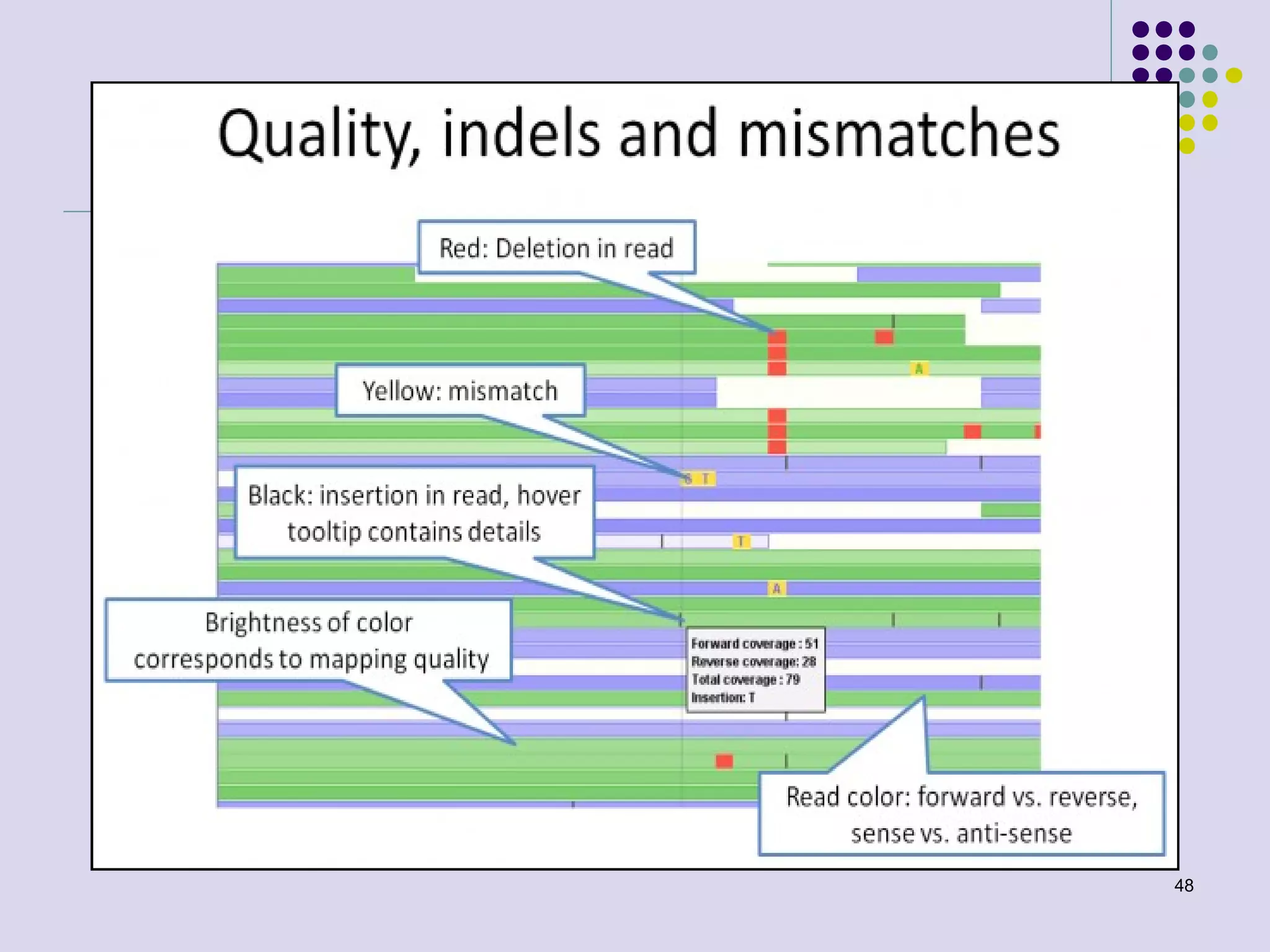
The document discusses genome annotation, covering its significance in biological understanding and the methodologies involved, including structural and functional genomics. It details the history and development of genome annotation techniques, highlighting the Human Genome Project and various gene-finding algorithms used for genomic analysis. The text emphasizes the importance of accurate annotation and the continuous evolution of gene predictions and functional assignments as new data emerges.