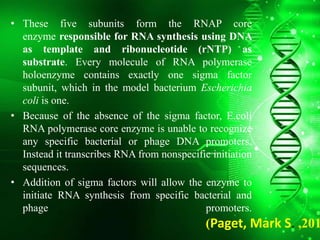
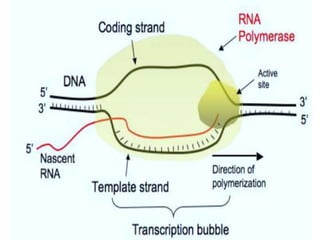
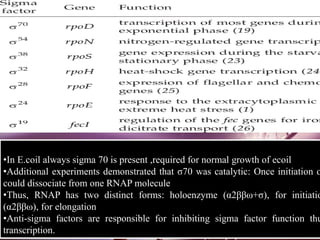
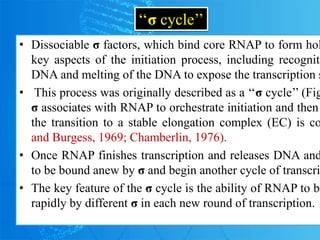
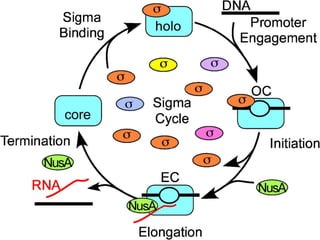
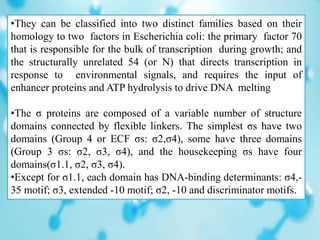
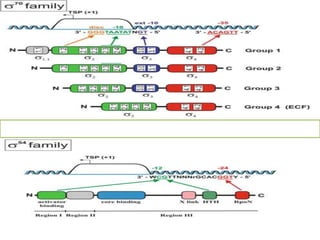
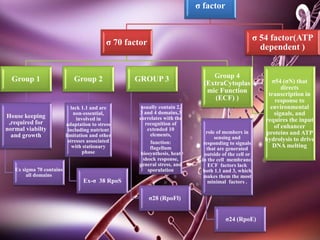
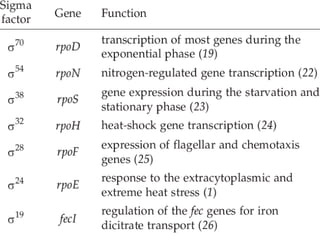
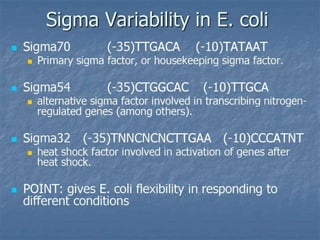
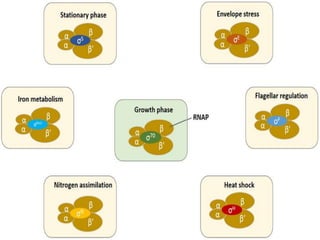
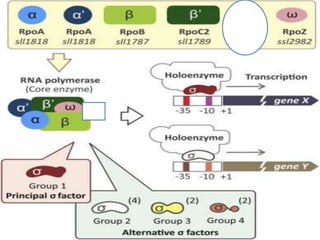
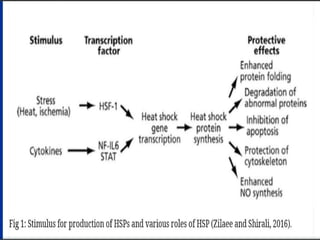
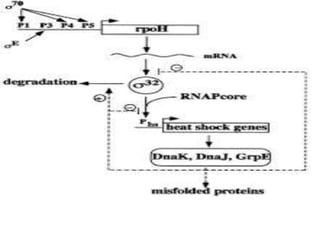
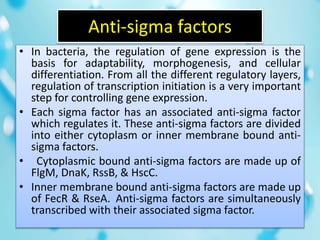
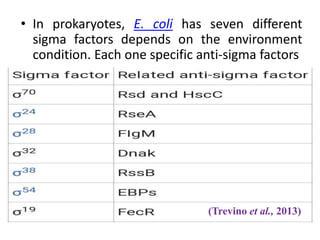
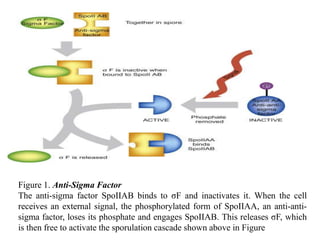
Sigma factors are subunits of bacterial RNA polymerase that play an important role in transcription initiation by recognizing promoter elements. E. coli contains seven sigma factors that direct transcription of different genes depending on environmental conditions. Sigma factors function by forming holoenzyme complexes with RNA polymerase core enzyme. Their activity is regulated by anti-sigma factors, which inhibit sigma factors and prevent transcription under certain conditions. During heat shock, sigma factor σ32 directs transcription of heat shock proteins by escaping inhibition of the anti-sigma factor DnaK.






![References
• Paget, Mark S. 2015. "Bacterial Sigma Factors and Anti-Sigma Factors: Structure,
Function and Distribution" Biomolecules 5, no. 3: 1245-1265.
https://doi.org/10.3390/biom5031245
• Saecker, R.M.; Record, M.T.; Dehaseth, P.L. Mechanism of bacterial transcription
initiation: RNA polymerase—Promoter binding, isomerization to initiation-
competent open complexes, and initiation of RNA synthesis. J. Mol. Biol. 2011,
412, 754–771.]
• Maria C. Davis, Christopher A. Kesthely, Emily A. Franklin, and Shawn R.
MacLellan. The essential activities of the bacterial sigma factor. Canadian Journal
of Microbiology. 63(2): 89-99. https://doi.org/10.1139/cjm-2016-0576
• Burgess RR, Travers AA, Dunn JJ, Bautz EK. Factor stimulating transcription by
RNA polymerase. Nature. 1969 Jan 4;221(5175):43-6. doi: 10.1038/221043a0.
PMID: 4882047.
• Treviño-Quintanilla LG, Freyre-González JA, Martínez-Flores I (September
2013). "Anti-Sigma Factors in E. coli: Common Regulatory Mechanisms Controlling
Sigma Factors Availability". Current Genomics. 14 (6): 378–
87. doi:10.2174/1389202911314060007. PMC 3861889. PMID 24396271](https://image.slidesharecdn.com/roleofsigmafactor-230418104048-7af6c978/85/role-of-sigma-factor-pptx-29-320.jpg)
