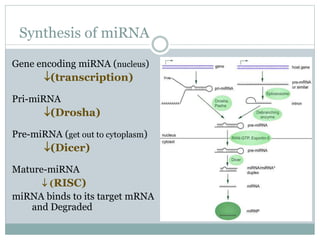
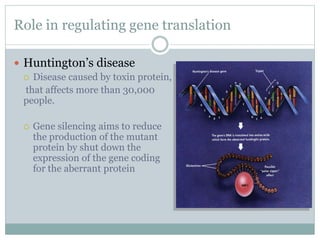
The document provides an overview of RNA interference (RNAi), detailing its mechanisms, discovery, and applications in gene silencing within eukaryotic organisms. It discusses the distinctions between small interfering RNAs (siRNAs) and microRNAs (miRNAs), their roles in regulating gene expression, and their implications in cancer pathogenesis and agricultural protection. RNAi is emphasized as a vital tool for controlling gene expression and addressing various biological challenges.