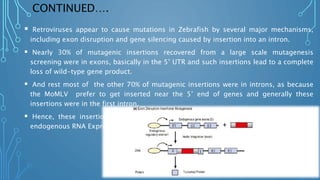
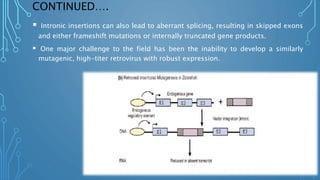
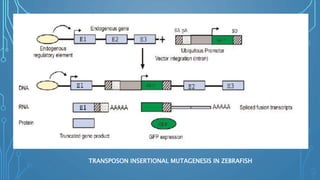
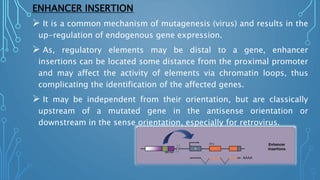
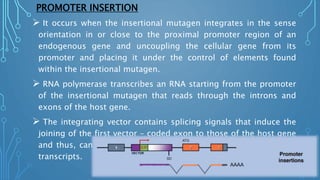
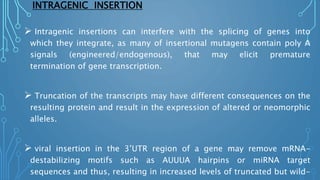
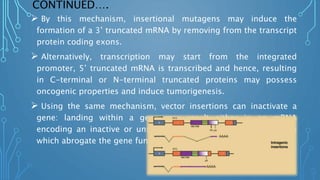
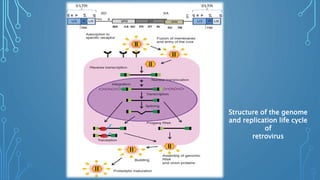
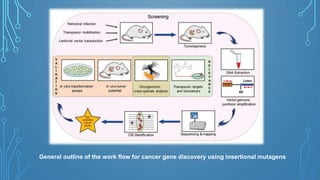
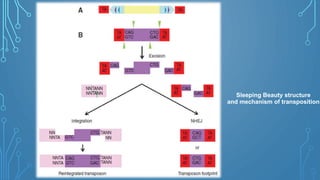
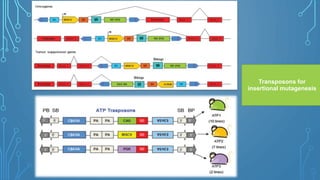
This document discusses insertional mutagenesis, specifically focusing on insertional mutagenesis. It defines insertional mutagenesis as the integration of exogenous DNA into a host genome, which can deregulate nearby genes and alter cellular phenotype. Retroviruses and transposons are commonly used as integrating agents in insertional mutagenesis experiments to identify novel cancer genes. The document describes how retroviruses like MoMLV and MMTV integrate randomly into the host genome and how analyzing common insertion sites from tumors can reveal cancer-causing genes. It also explains the different mechanisms of insertional mutagenesis, such as enhancer insertion, promoter insertion, and intragenic insertion, and how each can alter gene expression
























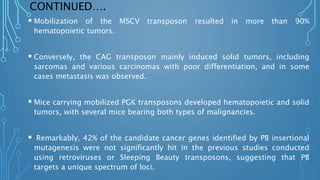




![CONTINUED….
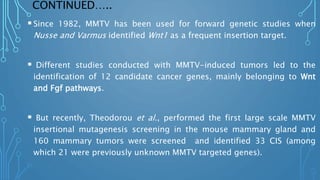
They established forward genetic tools for the zebrafish include chemical
(N-ethyl-N-nitrosourea [ENU]; and insertional (retroviral) mutagens.
Here, ENU produced random point mutations in the germline and these
single base pair changes resulted in a high frequency of mutant phenotypes.
In spite of the high efficiency in generation of point mutations, the major
limitation in this approach, was identification of genes whose mutations are
responsible for the particular phenotype.
An alternative approach is insertional mutagenesis, in which an exogenous
DNA serves as a mutagen and also functions as a molecular tag for
identifying the gene whose disruption causes the phenotype.](https://image.slidesharecdn.com/fbt-604-221002072430-617dc101/85/insertional-mutagenesis-39-320.jpg)