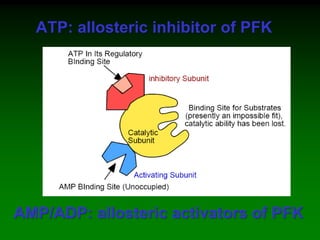
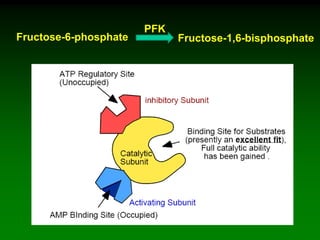
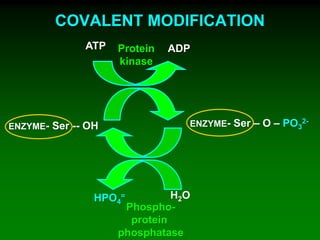
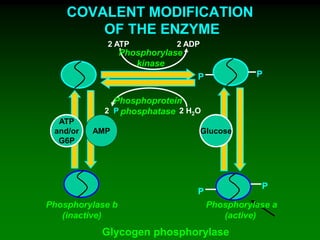
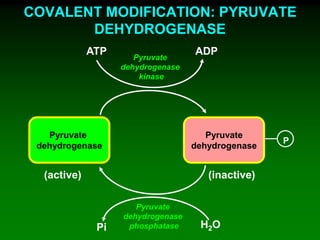
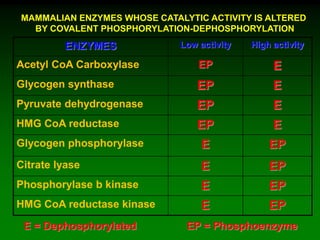
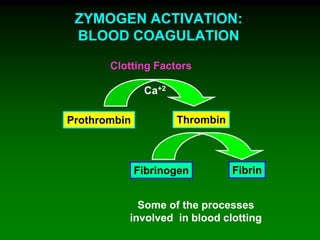
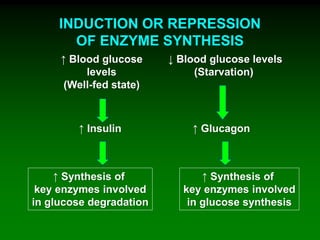
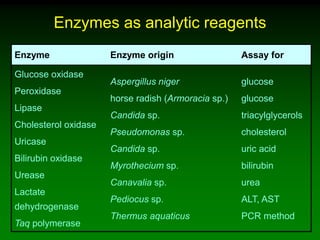
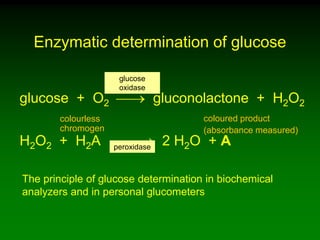
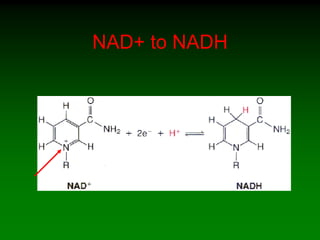
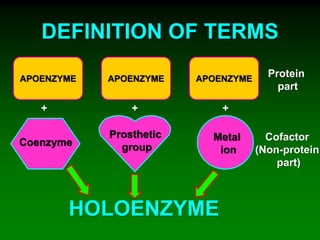
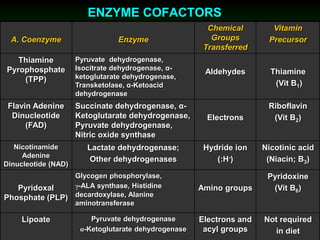
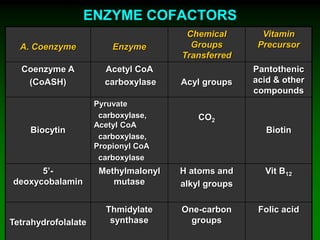
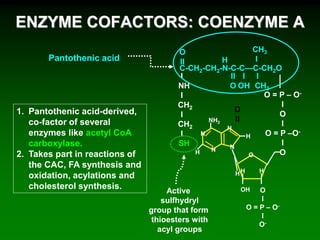
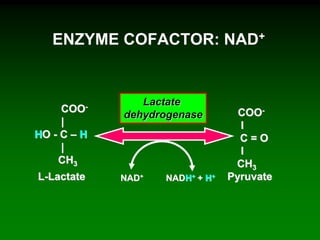
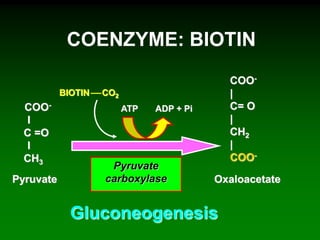
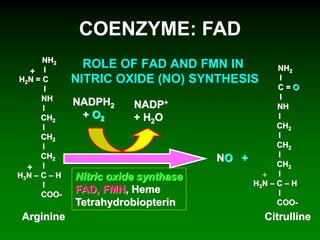
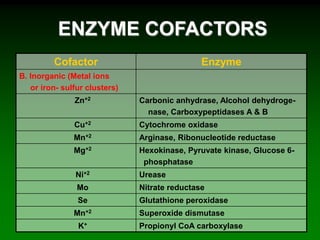
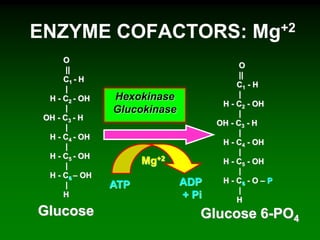
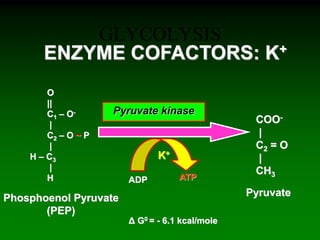
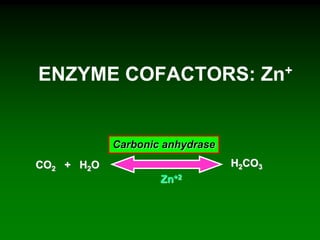
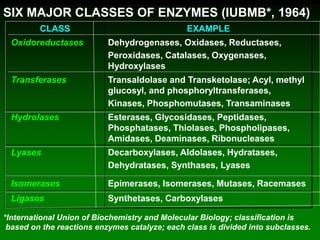
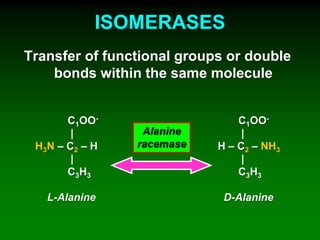
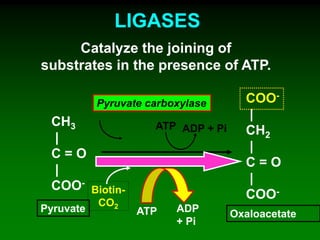
1) Enzymes require cofactors like vitamins and metal ions to function properly and carry out reactions. Common coenzymes include NAD+, FAD, coenzyme A, biotin, and vitamin B12.
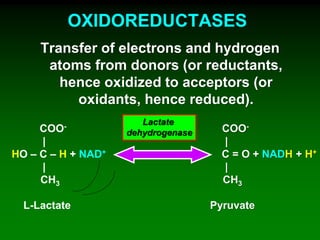
2) NAD+ acts as an electron acceptor in many oxidation reactions, FAD carries electrons and hydrogen atoms, and coenzyme A transfers acyl groups.
3) The cofactors help enzymes by participating directly in chemical transformations and bringing reactive groups close together to reduce the activation energy of reactions. Living systems rely on cofactors to drive essential metabolic pathways.




![Rate Constant: k
• A B
• Velocity of Rx
– V=d[B]/dt
• d=change
– V=-d[A]/dt
• V=d[B]/dt = -d[A]/dt = k[A]
• Large k rapid Rx
• Small k slow Rx](https://image.slidesharecdn.com/enzkinetics2014-140624211634-phpapp02/85/Enzkinetics-2014-11-320.jpg)

![1) Velocity is dependent on [S].
2) The more enzyme added, the
faster the reaction goes.](https://image.slidesharecdn.com/enzkinetics2014-140624211634-phpapp02/85/Enzkinetics-2014-15-320.jpg)
![Initial velocity Vo
• When enzyme is mixed with high
concentration of substrate [S] reaction goes
rapidly to steady state.
– Does not allow characterization
• Use low starting [S] and increase
• Hold [enzyme] constant
• Measure rate of reaction, Vo as [S] increases
– Until rate becomes constant: approaches Vmax](https://image.slidesharecdn.com/enzkinetics2014-140624211634-phpapp02/85/Enzkinetics-2014-16-320.jpg)
![Effect of [Substrate]](https://image.slidesharecdn.com/enzkinetics2014-140624211634-phpapp02/85/Enzkinetics-2014-17-320.jpg)
![A -------------> B
k1
A + B -----------> C
k2
d[A]/dt = k1[A]
For a first order reaction:
For a second order reaction:
The rate equation is:
The rate equation is:
d[A]/dt = k2[A][B]](https://image.slidesharecdn.com/enzkinetics2014-140624211634-phpapp02/85/Enzkinetics-2014-18-320.jpg)
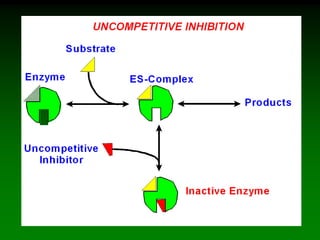
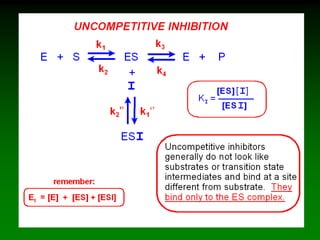
![E + S <=====> ES <=====> P + E
k2
k-2k-1
k1
In an enzymatic reaction:
But:
[ES] cannot be measured!
Nonetheless:
Initial [S] is known
[P] can be measured
[E]tot is known
Can we find [ES]?
Note: on initial conditions
[P] is negligibly small, So,
k-2[P]=0
Thus,
Vo=k2[ES] (1)](https://image.slidesharecdn.com/enzkinetics2014-140624211634-phpapp02/85/Enzkinetics-2014-19-320.jpg)
![YES!
If we assume "steady state kinetic conditions". That is, [S] and [P] are
changing, but [ES] does not change (a constant flux of S "through" the
enzyme).
d[ES]/dt = 0 (1)
Also (from conservation of matter):
[E]tot = [E]free + [ES] (2)
Now, divide eq. 1 by [E]tot :
Vo/[E]tot = k2[ES]/[E]tot
Since d[ES]/dt = 0 the rate of formation of [ES] must equal the rate of
breakdown of [ES]
Vformation = k1[E]free[S] (2nd order rate equation)
Vbreakdown= k2[ES] + k-1[ES]
= (k2 + k-1)[ES] (Two 1st order rate equations)
k1[E]free[S] = (k2 + k-1)[ES] (Rates must be equal)
E + S <=====> ES <=====> P + E
k2
k-2k-1
k1](https://image.slidesharecdn.com/enzkinetics2014-140624211634-phpapp02/85/Enzkinetics-2014-20-320.jpg)
![Rearranging, solving for [ES]:
[ES] = k1[E]free[S]/(k2 + k-1) (3)
If we define the Michaelis-Menten constant as Km = (k2 + k-1)/k1
and we substitute into eq. 3, we get:
[ES] = [E][S]/Km (4)
Now, let´s rearrange eq. 2 to [E]free = [E]tot - [ES] and then substitute into eq. 4
giving:
[ES] = ([E]tot - [ES])[S]/Km
Solving for [ES] gives:
[ES] = [E]tot(([S]/Km)/(1 + [S]/Km))
and then multiply top and bottom by Km, we get:
[ES] = [E]tot([S]/[S] + Km)](https://image.slidesharecdn.com/enzkinetics2014-140624211634-phpapp02/85/Enzkinetics-2014-21-320.jpg)
![[ES] = [E]tot([S]/[S] + Km)
Finally, substitute into eq. 1 to get:
Vo = k2[E]tot([S]/[S] + Km) (5)
this is the expression for Vo in terms of known quantities, let’s try it!!](https://image.slidesharecdn.com/enzkinetics2014-140624211634-phpapp02/85/Enzkinetics-2014-22-320.jpg)
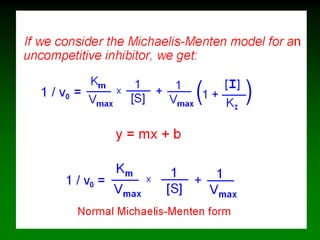
![If [S] very large
[S] >> Km. The enzyme is saturated with substrate.
In this case, [S] + Km = [S], so eq. 5 becomes:
Vo = k2[E]tot([S]/[S]) or Vo = k2[E]tot
This is the rate at large substrate concentration, the maximal rate for
[E], which is called Vmax
Vmax = k2[E]tot
Substituting into eq. 5 gives the Michaelis-Menten Equation:
Vo = Vmax[S]/Km + [S] (6)
Note: This equation is derived from Vo, very little [P] has formed.
Vo = k2[E]tot([S]/[S] + Km)](https://image.slidesharecdn.com/enzkinetics2014-140624211634-phpapp02/85/Enzkinetics-2014-23-320.jpg)
![If [S] is small:
[S] << Km Then the activity is in linear range.
[S] + Km = Km so eq. 6 becomes:
Vo = Vmax[S]/Km or Vo α [S]
Vo = k2[E]tot([S]/[S] + Km)
If [S] = Km
The definition of Km
Vo = Vmax[S]/[S] + [S] or
Vo = Vmax/2 = Km
Km is defined as the [S] that results in half maximal rate](https://image.slidesharecdn.com/enzkinetics2014-140624211634-phpapp02/85/Enzkinetics-2014-24-320.jpg)
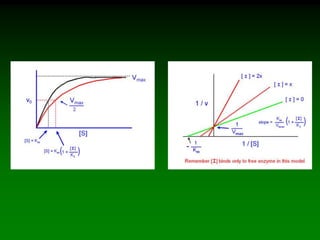
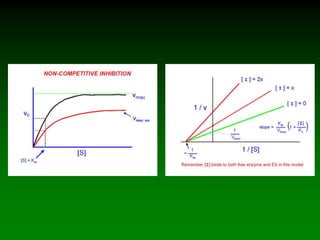
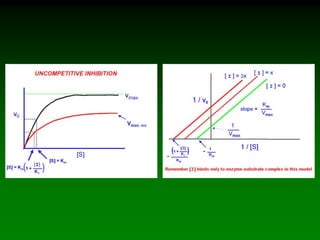
![Km and Vmax are
experimentally determined for
each enzyme using Vo vs. [S]
plots.
As V max is difficult to
determine (hyperbolic) the
Lineweaver-Burke plot is used.
Rearrange eq. 6 into linear form:
(1/Vo) = Km/Vmax(1/[S]) + 1/Vmax
So the plotted data looks like:](https://image.slidesharecdn.com/enzkinetics2014-140624211634-phpapp02/85/Enzkinetics-2014-25-320.jpg)


![Derivation of KCAT or turnover number:
For all catalyzed reactions:
[E]tot = [E]free + [ES] (2)
And with [S] >> Km:
Vmax = k2[E]tot or
k2 = Vmax/ [E]tot
For this reason k2 is also known as KCAT when the enzyme is saturated, so:
KCAT = Vmax/ [E]tot (7)
Therefore, when [S] is low, from eq. 7 and the Michaelis-Menten equation:
V = Vmax[S]/Km
then
V = KCAT/Km[S][E]
Therefore, KCAT/Km is a measure of how
rapidly and enzyme works and is referred as
the specificity constant or catalytic efficiency](https://image.slidesharecdn.com/enzkinetics2014-140624211634-phpapp02/85/Enzkinetics-2014-28-320.jpg)



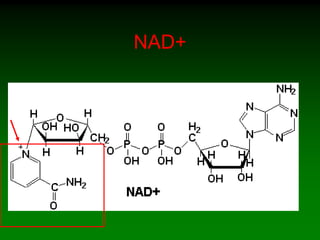
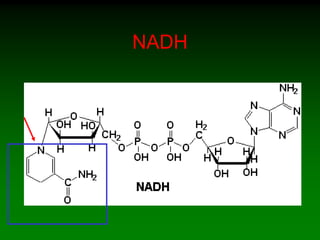


![WHAT ARE WE MEASURING ?
• Production of NADH
– NAD+ NADH
– Wavelength shift
• Depends on participation of Alcohol and ADH
• How can you do this
• Ensure that NAD is not a rate limiting
component.
– [NAD] constant and high
– [ADH] constant
– [ETOH] low and increasing](https://image.slidesharecdn.com/enzkinetics2014-140624211634-phpapp02/85/Enzkinetics-2014-38-320.jpg)
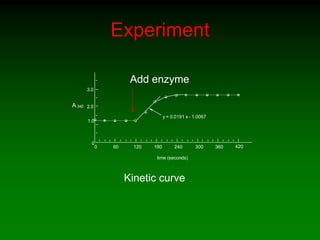
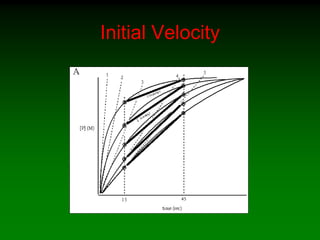
![Effect of enzyme concentration
I
II
[E]std
[P]
time (min)
0 2 4 6 8 10](https://image.slidesharecdn.com/enzkinetics2014-140624211634-phpapp02/85/Enzkinetics-2014-42-320.jpg)



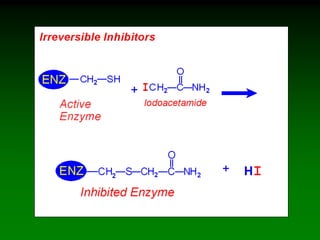
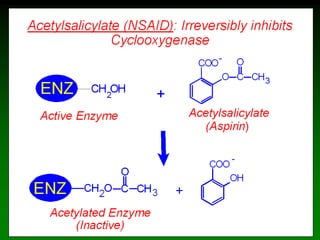

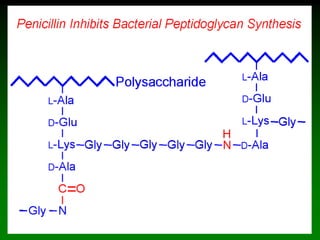
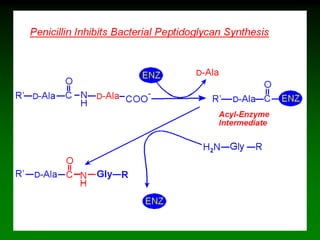
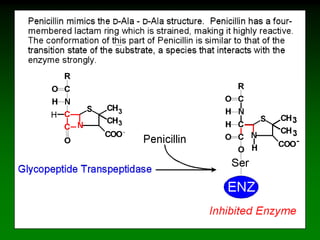

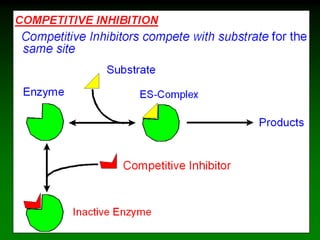
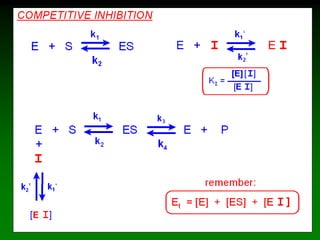
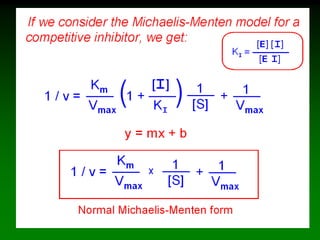
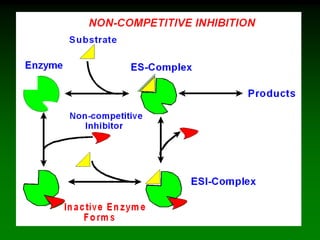
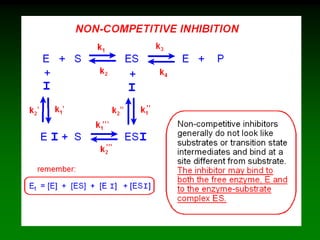
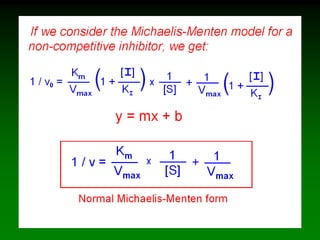


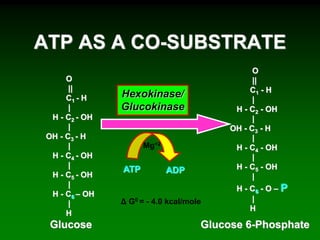



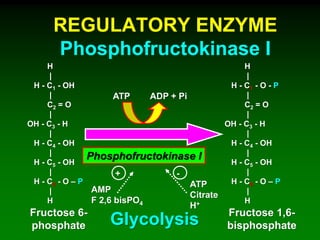

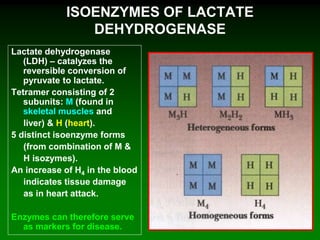


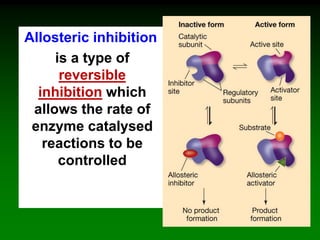
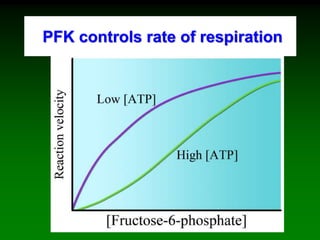
![98
Saturation curve of allosteric enzymes is sigmoidal
[S]
vo aktivace
inhibicebez
efektoru
activation
without
effector
inhibition](https://image.slidesharecdn.com/enzkinetics2014-140624211634-phpapp02/85/Enzkinetics-2014-98-320.jpg)

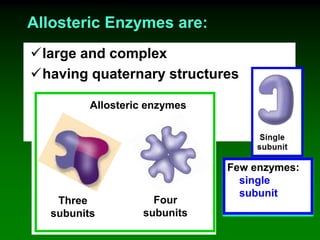
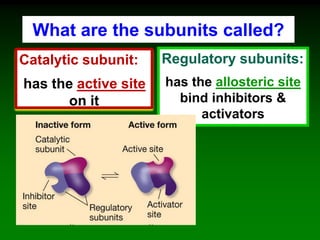
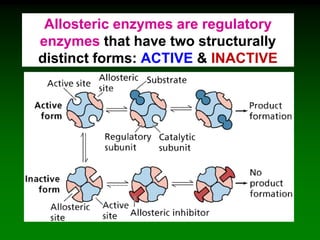
![‘Allostery’ means ‘different
shape’ [refers to the two shapes
of the enzyme]
Allosteric enzymes have TWO sites:
E
Active site
Allosteric site
Substrate
cannot fit into
the active site
Inhibitor
molecule
Inhibitor fits into
allosteric site
E](https://image.slidesharecdn.com/enzkinetics2014-140624211634-phpapp02/85/Enzkinetics-2014-102-320.jpg)